BIGNASim database structure and analysis portal for nucleic acids simulation data
----
DNA & RNA Structure and Helical Parameters Analyses (NAFlex)
----Tutorial 2 -- Global analysis (XCGY)
Tutorial 3 -- Meta-trajectory (XCGY)
Tutorial 4 -- Experimental vs MD analysis
----Searching
The easiest way to search in BIGNASim server is using the Quick Search placed in the right part of the main menu tab. This form allows searching by PDB Id, ontology terms, keywords and authors.

In addition to the Quick Search, BIGNASim portal offers a powerful searching engine to find particular sequences/simulations. Basically, the search can be divided into three main sections:
- By sequence or specific sequence fragments (using regular expressions)
- By specific base-pair-steps (with or without flanking regions)
- By an extensive nucleic acid ontology (as defined here)
Once the search is done, a browse page will appear showing the resulting trajectories.
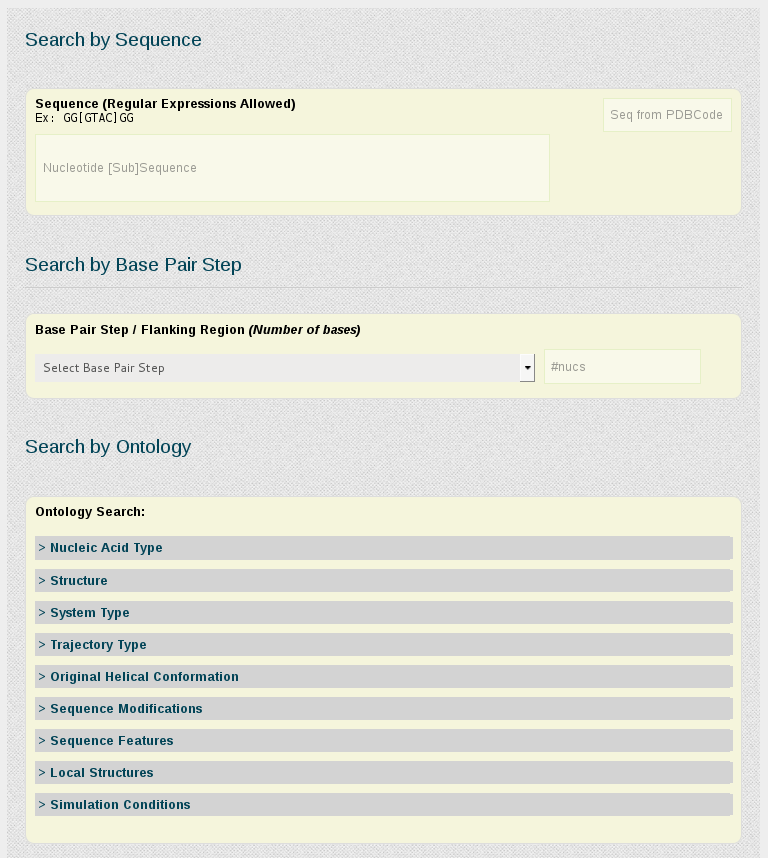
Search by Sequence

The search engine is able to find a particular sequence, with the possibility to use regular expressions, which increases the power of the search. Input needed is just a chain of bases [A,C,T,G,U]. Examples:
- Sequence (DDD): CGCGAATTCGCG
- Sequence Fragment: CTAG
Examples of interesting regular expressions could be:
- Sequence fragment with a variable nucleotide: GG[GTAC]GG
- Sequence fragment with flanking region: [ACGT]{2}CTAG[ACGT]{2}
- Repetition of bases: [G]{4}
- Sequences beggining/ending with a particular fragment: ^AA / AA$
Nucleic Acid sequence associated to a particular PDB file will be recovered if a PDB code is used as input in the Seq from PDB Code box. This allows to locate also other PDB entries sharing the same sequence or with small modifications.
Search by Base-Pair Step
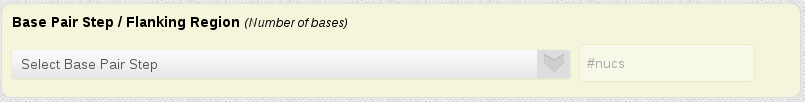
One of the most important study to understand nucleic acids flexibility is applied in a base-pair step region. This section of the search engine allows to easily select a particular base-pair step to find all sequences simulated having this particular pair of nucleotides.
Typically, base-pair step studies omit terminal bases, to avoid fraying effects. For this reason, a flanking region (number of bases) can be given, and only those base-pair step having this number of bases before and after them will be retrieved.
Search by Ontology

All simulations stored in BIGNASim system are classified using a Nucleic Acids Ontology (see Ontology section).
The search engine is querying the database system on the fly, and showing the number of results found in the top-right corner of the ontology search box.
The ontology is split in 8 different sections:
- Nucleic Acid Type:
 Nucleic Acid type: DNA, RNA or DNA-RNA Hybrid.
Nucleic Acid type: DNA, RNA or DNA-RNA Hybrid.
- Structure:
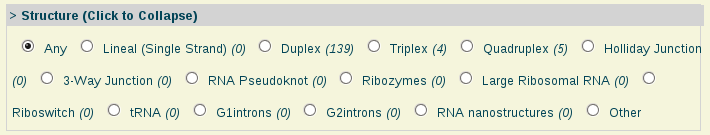 Structure: Single-stranded nucleic acid, duplex, triplex, quadruplex and other nucleic acids structure types.
Structure: Single-stranded nucleic acid, duplex, triplex, quadruplex and other nucleic acids structure types.
- System Type:
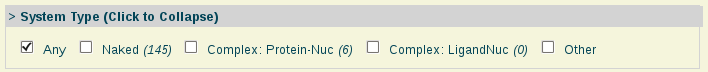 System type: Nucleic acid naked or complexed with other molecules.
System type: Nucleic acid naked or complexed with other molecules.
- Trajectory Type:
 Trajectory Type: Nucleic acid trajectory computed in equilibrium, folding/unfolding or transition simulation.
Trajectory Type: Nucleic acid trajectory computed in equilibrium, folding/unfolding or transition simulation.
- Original Helical Conformation:
 Original Helical Conformation: Nucleic Acid original helical conformation: A, B, Z, Hoogsteen, mixed (transitions), etc.
Original Helical Conformation: Nucleic Acid original helical conformation: A, B, Z, Hoogsteen, mixed (transitions), etc.
- Sequence Modifications:
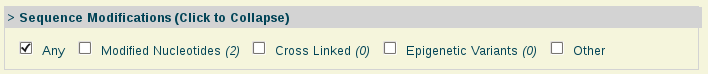 Sequence Modifications: Nucleic acids with modified nucleotides.
Sequence Modifications: Nucleic acids with modified nucleotides.
- Sequence Features:
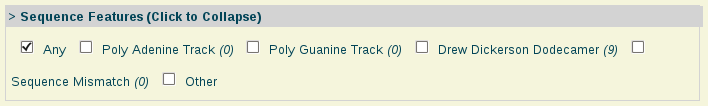 Sequence Features: Nucleic acids with special sequence features such as the well-known Drew-Dickerson Dodecamer (DDD), sequences having mismatches, etc.
Sequence Features: Nucleic acids with special sequence features such as the well-known Drew-Dickerson Dodecamer (DDD), sequences having mismatches, etc.
- Local Structures:
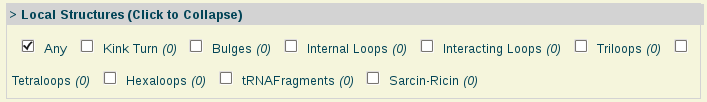 Local Structures: Different types of local structures arrangements, specially related to RNA molecules.
Local Structures: Different types of local structures arrangements, specially related to RNA molecules.
- Simulation Conditions:
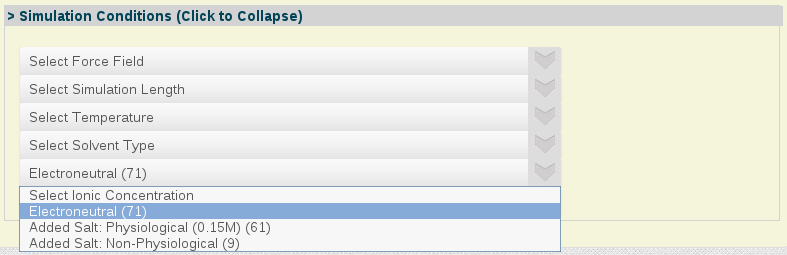 Simulation Conditions: A selected set of simulation condition parameters, from force-field used to system ionic concentration.
Simulation Conditions: A selected set of simulation condition parameters, from force-field used to system ionic concentration.



