MC DNA Help - Analysis Contacts
Distance Contact Maps
Method
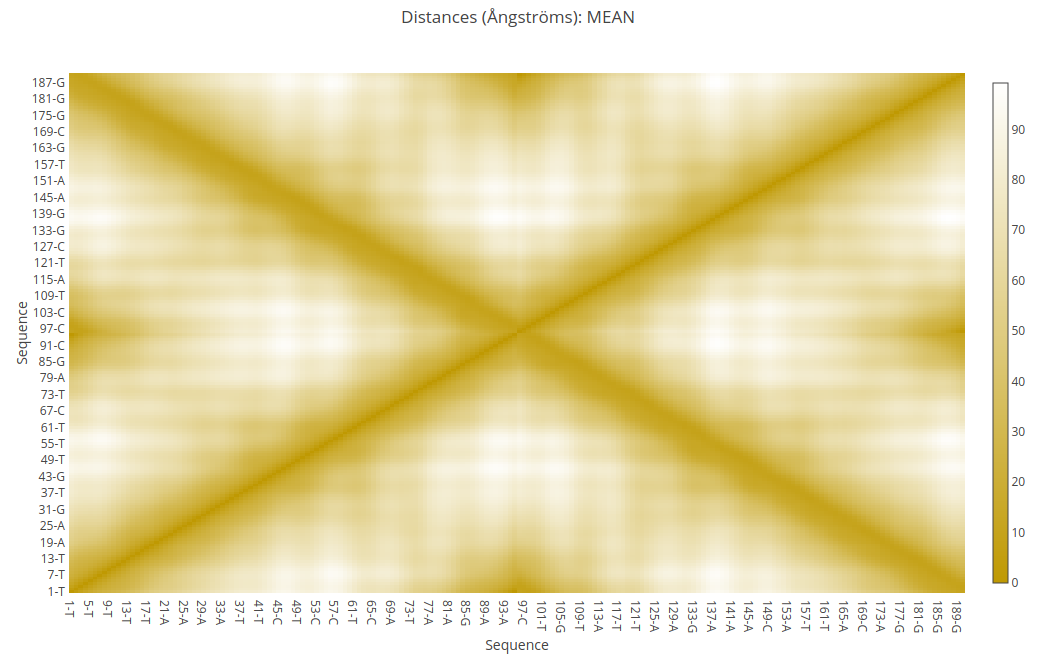
Information as the distances (for example distance mean or minimum distance) between all the pair of bases in a nucleic structure during a dynamic simulation can be very useful to obtain a picture of the flexibility of the structure and the movements that it undergoes during the dynamic simulation.
The most common way to look at this information is plotting it with Contact Maps. With these plots, a square is painted for every nucleotide pair, assigning a color depending on the value of the analysis (distance in this case). Usually, high values out of the diagonal (closest nucleotide pairs) are the most interesting and informative ones.
Results
In this analysis, user just have to choose between the different Distance Contact Maps offered by the server clicking at the corresponding button. The plot will be shown with a color legend, and like all the different plots generated by MC DNA, with the associated links giving the possibility to open it in a new window and download the raw data.
Nucleotide - Nucleotide
Distances for all pairs of nucleotides (all against all) in the sequence are computed in all methods offered by MCDNA server. When the type of simulation chosen is a structure generation, a unique plot will be shown, using the distances found analysing the final structure. When the type of simulation chosen is a trajectory (ensemble) generation, 4 different plots will be presented, using the distances found analysing all the snapshots in the ensemble. The four plots are: Mean (distances mean along the ensemble), Min (minimum distance found for the particular nucleotide pair in all the structures of the ensemble), Max (maximum distance found for the particular nucleotide pair in all the structures of the ensemble), and Stdev (distance standard deviation found along the ensemble).

Protein - Nucleotide
Results for the MC-DNA + Proteins tool generate two specific sub-sections in the Contacts analysis: Protein-Nucleotide and Protein-Protein distances. In these cases, one (for structure) or four (for trajectory/ensemble) different plots are computed and showed in each of the sub-sections when a protein (or two proteins in the Protein-Protein case) is selected.
Distances shown in Protein-Nucleotide section correspond to the distances between the selected protein and the nucleotide
To view these plots, user first have to select the desired protein from a list of all the proteins selected for the simulation in the input section of the server. To help the user, a 3D view of the nucleic structure together with the the proteins is shown using NGL viewer.

After selecting the protein and clicking the Load contacts button, the heatmap plot corresponding to the distances between the protein and the nucleotides will be shown.

Protein - Protein
RESULTS PROT - PROT EXPLANATION
Results for the MC-DNA + Proteins tool generate two specific sub-sections in the Contacts analysis: Protein-Nucleotide and Protein-Protein distances. In these cases, one (for structure) or four (for trajectory/ensemble) different plots are computed and showed in each of the sub-sections when a protein (or two proteins in the Protein-Protein case) is selected.
Distances shown in Protein-Nucleotide section correspond to the distances between the selected protein and the nucleotide
To view these plots, user first have to select the two desired proteins from a list of all the proteins selected for the simulation in the input section of the server. To help the user, a 3D view of the nucleic structure together with the proteins is shown using NGL viewer.

After selecting the proteins and clicking the Load contacts button, the heatmap plot corresponding to the distances between both proteins will be shown.

