BioMoby Ontology
MDWeb is internally powered by BioMoby Services and Ontology.
 BioMoby (www.biomoby.org) defines an ontology-based messaging standard through which a client will be able to
automatically discover and interact with task-appropriate biological data and service providers, without requiring manual manipulation
of data formats as data flows from one provider to the next.
BioMoby (www.biomoby.org) defines an ontology-based messaging standard through which a client will be able to
automatically discover and interact with task-appropriate biological data and service providers, without requiring manual manipulation
of data formats as data flows from one provider to the next.
The use of such object ontology allows to have:
- Service Inputs/Outputs with biological meaning.
- Service automatic discovery, through possible input objects.
In MDWeb project, a complete Molecular Dynamics ontology has been built embracing the most popular MD programs and files formats, such as Gromacs, Amber and NAMD. Due to the lack of a standard for the definition of MD data the present implementation, although offering a uniform framework, keeps the native low-level implementation of each MD package.
BioMoby Molecular Dynamics Ontology has a hierarchical organization. All objects inherits from a parent object called: Object.
In this help section a complete Molecular Dynamics Ontology definition can be found, object by object, as well as a set of graphical diagrams.
Ontology Diagrams
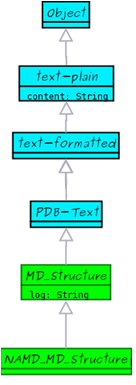
Ontology Objects
In the following, object relationships as expressed as: IS_A: simple inheritance, HAS_A: contained field, HAS: contained collection of fields.
- Base Objects
- Objects including data in native text formats
- Molecular Dynamics Topology Data
- Molecular Dynamics Structure Objects
- Molecular Dynamics Trajectory Objects
- Molecular Dynamics Restart Objects
- MD Configuration Files
Base Objects
Object |
| BioMoby Base Object. This is the base object of any BioMoby ontology. Object contains only an Id and a Namespace where the Id has a meaning (ex. 1ubq in PDB) |
Text-plain |
Object containing a plain text.
|
Text-formatted |
Object containing a text, following a standard bioinformatics format (PDB, FASTA, etc.). All common bioinformatics text-based formats should inherit from this object.
|
Objects including data in native text formats
PDB-Text |
Structure in PDB format.
|
GROMACS_Gro_Text |
A molecular structure in Gromacs Gromos87 format.
|
GROMACS_ITP_Text |
Molecular Dynamics Topology Include Files in Gromacs Format. GROMACS ITP files (to be included in GROMACS topology) describing the molecular structures.
|
NAMD_Velocities |
NAMD Molecular Dynamics Restart Velocities file. NAMD Restart File storing the current Velocities of all atoms at some step of the simulation.
|
NAMD_XSC |
NAMD Molecular Dynamics Restart XSC file. XSC eXtended System Configuration file, containing a record of the periodic cell parameters and extended system variables.
|
NAMD_Positions |
NAMD Molecular Dynamics Restart Positions file.NAMD Restart File storing the current positions of all atoms at some step of the simulation.
|
NAMD_Velocities |
NAMD Molecular Dynamics Restart Velocities file. NAMD Restart File storing the current Velocities of all atoms at some step of the simulation.
|
NAMD_XSC |
NAMD Molecular Dynamics Restart XSC file. XSC eXtended System Configuration file, containing a record of the periodic cell parameters and extended system variables.
|
NAMD_Positions |
NAMD Molecular Dynamics Restart Positions file. NAMD Restart File storing the current positions of all atoms at some step of the simulation.
|
AMBER_Lib |
Ligand Library in AMBER format.
|
AMBER_Frcmod |
Ligand Frcmod in AMBER format.
|
Molecular Dynamics Topology Data
MD_Topology |
|
Molecular Dynamics Topology. Topologies define the nature of the simulated system, including force-field parameters, atom/residues description and connectivities. MD_Topology is defined as generic. Descendants will determine the format used (Gromacs, NAMD, Amber), and include the necessary additional fields.
|
|
| Specific Topology objects | |
GROMACS_TOP_Text |
|
Molecular Dynamics Topology in Gromacs Format. GROMACS format TOP file describing the molecular structure.
Additional fields are the topology, and a collection of related itp parameter sets.
|
|
PSF_Text |
|
Molecular Dynamics Topology in PSF Format. X-Plor format PSF file describing the molecular structure.
|
|
PRMTOP_Text |
|
Molecular Dynamics Topology in PRMTOP Format. PRMTOP Format Topology file describing the molecular structure.
|
|
Molecular Dynamics Structure Objects
MD_Structure |
|
| Molecular Dynamics Structure. It contains a structure in PDB format and a log report of the program that has generated the structure. Used to store structures that have been already prepared for simulation.
MD_Structure objects are defined as generic. Descendants will determine the precise format used (Gromacs, NAMD, Amber), and the necessary additional data fields.
|
|
| Specific MD_Structure objects. | |
GROMACS_MD_Structure |
|
Molecular Dynamics Structure in Gromacs Format. MD_Structure prepared from simulation in GROMACS. Addiitional fields are a topology in Gromacs TOP format, and a structure in Gromacs GRO format.
|
|
NAMD_MD_Structure |
|
Molecular Dynamics Structure in NAMD Format.Structure prepared from simulation in NAMD. Additional field is a topology in X-Plor's PSF.
|
|
AMBER_MD_Structure |
|
Molecular Dynamics Structure in Amber Format.Structure prepared from simulation in AMBER. Additional field is a topology in PRMTOP format.
|
|
Molecular Dynamics Trajectory Objects
MD_Trajectory |
|
Molecular Dynamics Trajectory. Molecular Dynamics Output Trajectory containing MD Topology, Coordinates and Restart Files.
MD_Trajectory is defeined a in generic form, descendants will determine the precise format used (CRD, Binpos, DCD, NetCDF, XTC).
Included MD_Structure, and MD_Restart objects should be format aware objects indicating the MD package used to generate the trajectory.
|
|
| Specific MD_Trajectory objects | |
MD_TrajectoryCRD |
|
CRD Molecular Dynamics Trajectory. Molecular Dynamics Output Trajectory containing MD Topology, Coordinates and Restart Files. ASCII CRD Format.
|
|
MD_TrajectoryBINPOS |
|
BINPOS Molecular Dynamics Trajectory. Molecular Dynamics Output Trajectory containing MD Topology, Coordinates and Restart Files. Scripps Binpos Binary Format.
|
|
MD_TrajectoryDCD |
|
DCD Molecular Dynamics Trajectory.Molecular Dynamics Output Trajectory containing MD Topology, Coordinates and Restart Files. Charmm DCD Binary Format.
|
|
MD_TrajectoryNetCDF |
|
NetCDF Molecular Dynamics Trajectory. Molecular Dynamics Output Trajectory containing MD Topology, Coordinates and Restart Files. NetCDF (Network Common Data Form), machine-independent binary data format for array-oriented scientific data.
|
|
MD_TrajectoryXTC |
|
XTC Molecular Dynamics Trajectory. Molecular Dynamics Output Trajectory containing MD Topology, Coordinates and Restart Files. Gromacs binary XTC Format.
|
|
MD_Compressed_Trajectory |
|
Compressed Molecular Dynamics Trajectory. MD Trajectory in compressed PCZ format (PCAZip Compressed Binary format), built with PCAsuite (Software package for lossy trajectory compression using Principal Component Analysis (PCA) techniques).
|
|
Molecular Dynamics Restart Objects
MD_Restart |
|
Molecular Dynamics Restart. Molecular Dynamics Restart Files (Coordinates, Velocities and Box info) that allows a dynamic simulation to continue from a certain point. Defined as generic object, descendants will determine the format used (Gromacs, Namd).
|
|
| Specific MD_Restart objects | |
NAMD_Restart |
|
NAMD Molecular Dynamics Restart. NAMD MD Restart File containing Velocities, Positions and Extended System Configuration.
|
|
GROMACS_Restart |
|
GROMACS Molecular Dynamics Restart. MD Restart File containing Gromacs EDR and TRR binary files.
|
|
GROMACS_TRR_Restart |
|
GROMACS Molecular Dynamics TRR Restart file. GROMACS Restart File (TRR) storing the current periodic box information, and Velocities and Positions of all atoms at some step of the simulation (Portable binary format).
|
|
GROMACS_EDR_Restart |
|
GROMACS Molecular Dynamics EDR Restart file. GROMACS Restart Energy File (EDR) storing energies, temperature, pressure, box size, density and virials (Portable binary file).
|
|
MD Configuration Files
GROMACS_Conf_Text |
GROMACS Configuration File.
|
NAMD_Conf_Text |
NAMD Scalable Molecular Dynamics Configuration File.
|