NAFlex Atomistic Molecular Dynamics Simulations
Since first simulation of biomacromolecules in 1977, molecular dynamics (MD) has experienced a long evolution,
and, at present, it is a mature technique that can be used to obtain an accurate picture of the dynamics of complex systems with atomistic detail.
Molecular Dynamics (MD) is a deterministic technique which allows predicting a system’s future states based on its actual state defined by positions and impulses of all
particles. Its foundations are based on the laws of classical mechanics which define the equations of motion used to generate successive configurations and thus progress the
system through time.

Newton’s second law of motion describes the acceleration of a particle i of mass mi along a coordinate xi.
The force Fi acting upon particle i depends on the positions of all other particles in the system which makes its potential energy surface extremely
complex and impedes direct integration. Instead, tedious numerical integration in small time steps are needed, making MD simulations computationally costly.
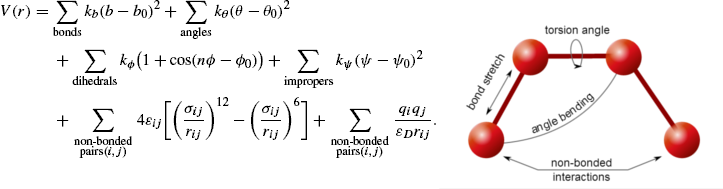
Forcefields link a structure’s atomic coordinates to a potential energy. Since every structure refers to one configuration in the proteins conformational
space the forcefield gives a complete description of the proteins potential energy surface.
Forcefields consist of two components, the Bonded terms and the Non-bonded terms. Bonded terms are those describing bond, angle, and dihedral interactions whereas Non-bonded terms account for distant interactions and describe electrostatic and Van der Waals interactions.
Despite all its power the practical use of MD is limited by three factors:
i) the uncertainties of classical force-fields.
ii) the required computational power.
iii) the high level of expertise needed to use the technique.
Focusing in the last point, it should be remarked that setting up a system for simulation requires from the combination of a series of operations,
many of them to be performed manually, and a number of decisions that demand a significant degree of expertise. The end result is that newcomers to the field face a stiff learning curve and that in some cases (for example when supercomputer
resources are used) setting-up of simulations can represent a period of time similar to that of deriving the trajectory. These problems reach a maximum in high-throughput projects, where thousands of trajectories need to be launched, what forces the development of automatic tools for mimicking human expertise in launching (and then analyzing) MD trajectories.
MDWeb: Molecular Dynamics on Web

Atomistic Molecular Dynamics Setup (preparation of the system to be launched) and Simulation in NAFlex are powered by our web server MDWeb and the web-services package for Molecular Dynamics MDMoby.
 Within the MoDEL project we have developed a series of tools that allowed us to launch and analyze
around 2,000 trajectories with little human intervention. An evolution of our original workflows is made available in our MDWeb server as a general tool,
not linked to any specific simulation package, to help naïve users to prepare completely systems for simulations,
and that allows expert users the use of MD in the high-throughput regime.
Within the MoDEL project we have developed a series of tools that allowed us to launch and analyze
around 2,000 trajectories with little human intervention. An evolution of our original workflows is made available in our MDWeb server as a general tool,
not linked to any specific simulation package, to help naïve users to prepare completely systems for simulations,
and that allows expert users the use of MD in the high-throughput regime.
The software tool developed in MDWeb is presented as a fully integrated web-services-oriented software platform to perform molecular dynamics (MDMoby), and its web portal MDWeb server.
The technology has been adapted to be accessed as web-services following the BioMoby framework.
MDMoby can be accessed with web-services clients and also programatically through suitable APIs (INB-BSC).
A number of pre-prepared pipelines are also available to help non-expert users in the use of standard simulation procedures.
The web portal MDWeb provides a friendly environment to setup new systems, run test simulations and perform analysis within a guided interface.
Setup files can be prepared for Amber, NAMD, and Gromacs (other formats will be incorporated in the future) and analysis can be carried out using any standard
trajectory format.
 Additionally, the platform is interfaced to our flexibility analysis software FlexServ, so providing coarse-grained
simulation, and advanced flexibility analysis tools for Proteins.
Additionally, the platform is interfaced to our flexibility analysis software FlexServ, so providing coarse-grained
simulation, and advanced flexibility analysis tools for Proteins.