Uploading a Trajectory in NAFlex
NAFlex provides a friendly environment to analyse your own generated molecular dynamics trajectories. With this short tutorial, you will be able to upload a trajectory and run a set of analysis, checking for example the stability of your system or information about flexibility.
Tutorial Steps
- Registration
- Starting Project
- Uploading a trajectory
- Running Analysis
- Trajectory manipulation: format conversions, slice/snapshot extraction, etc.
- Analysis per Residue/Structure.
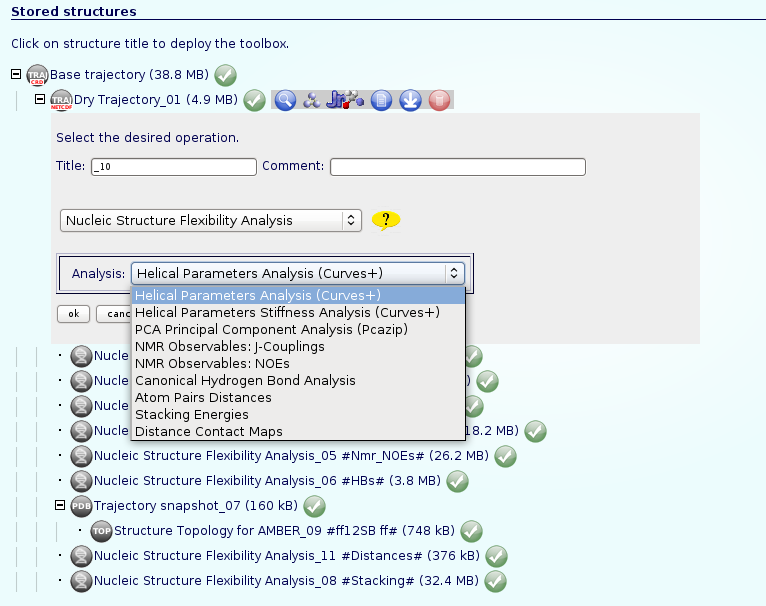
- Nucleic Acids Flexibility Analysis.
The first thing to do is to choose between working as an anonymous user or alternatively as a registered user. We strongly recommend working as a registered user, as it has some important advantages.
Anonymous user's projects are completely removed once the user is disconnected and also when session expires (after some minutes of inactivity), and therefore working as anonymous user is only suited for a first impression of the web server.
Registration process will just take a minute --> Registration.
Once logged in, the user workspace appears. In this workspace, all projects of the user will be shown.
Now, we are ready to start our first NAFlex analysis project.

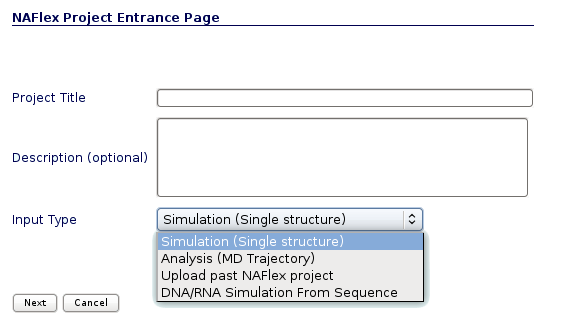
NAFlex user can choose between four different kinds of inputs, Simulation, Analysis, DNA/RNA Simulation from Sequence and Upload a past NAFlex project. Now we will see an example of an Analysis project.
You will be asked for a small set of data and input files: First of all, you may fill in a project title and a description (optional) at the top part of the form. Then, at the Input Type drop-down selector, just under the description, you should choose the option Analysis (MD Trajectory).
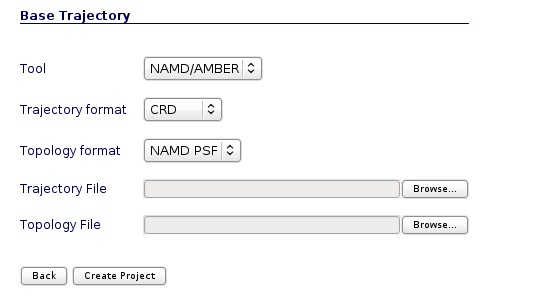
User may select the appropiate tool, trajectory and topology format to work with the input files. In the next section we will see an example of how to build the trajectory and topology files with a size small enough to be transfered through the net to NAFlex server.
Molecular dynamics generated trajectories are usually very big files, containing all the water molecules and counterions included in the solvent box, and with a fine resolution such as a snapshot for each picosecond. NAFlex is an on-line server and thus it has the limitations regarding the transfer of big amounts of data through the net, forcing us to limit the trajectory size to 100MB.
So, more than likely, first of all we will need to reduce the size of the MD-generated trajectory to be uploaded to NAFlex. We can just extract a representative set of snapshots from the original obtained trajectory, (for example one snapshot every 0.5ns). Alternatively, if we wish to have more resolution, we can extract more snapshots by removing solvent molecules, thus drastically reducing the trajectory file size, but to be able to properly analyse this solvent-stripped trajectory, a coherent topology must be generated, and that's usually not an easy job.
Example of reducing trajectory size (click to expand):
XTC Trajectory
Use trjconv from Gromacs package to reduce the size of a trajectory in XTC format.
Usage: trjconv_sp -s structure.gro -f trajectory.xtc -o trajectory.20.xtc -b 1 -e 10000 -dt 500 -ur compact -pbc atom
where structure.gro is the structure topology in GRO format, trajectory.xtc is the trajectory in XTC format, 1-10000 are the starting and ending snapshot times (in ps), and 500 is the offset (write every 500 ps). The rest of the commands are useful when dealing with non-cubic boxes (-ur) and periodic boundary conditions (-pbc).
You will be asked for a group to write to the output file, e.g.:
Will write xtc: Compressed trajectory (portable xdr format)
Select group for output
Opening library file /usr/local/gromacs/share/gromacs/top/aminoacids.dat
Group 0 ( System) has 99637 elements
Group 1 ( Protein) has 8380 elements
Group 2 ( Protein-H) has 4263 elements
Group 3 ( C-alpha) has 534 elements
Group 4 ( Backbone) has 1602 elements
etc.
We are going to choose System (group 0), to write all the system as output and hence being coherent with the system topology.
Executing these commands will generate a new reduced trajectory, named trajectory.20.xtc, in Gromacs XTC format, ready to be uploaded to NAFlex server.
Use ptraj from Ambertools package to reduce the size of a trajectory in DCD, NetCDF, Binpos or CRD formats.
Usage: ptraj structure.top < ptrajReducing.in
where structure.top is the structure topology in PRMtop or PSF format, and ptrajReducing.in is a file containing the apropriate commands:
trajin trajectory.crd 1 10000 500 # Input trajectory, first and last snapshot to read and offset.
trajout trajectory.20.crd # Output trajectory, in Amber ASCII CRD format (ptraj default).
go
Executing these commands will generate a new reduced trajectory, named trajectory.20.crd, in Amber ASCII CRD format, ready to be uploaded to NAFlex server.
To reduce a little bit more the generated trajectory, it can be compressed with gzip program (as it is an ASCII file). NAFlex is able to deal with this kind of compressed files.
Alternatively, trajectory can be generated in binary netCDF format, thus reducing a lot the final file size, but for ptraj to be able to generate this kind of file, it needs to be compiled with the corresponding netCDF libraries.
To generate the final trajectory in binary netCDF format, replace the trajout command line for this new one:
trajout trajectory.20.netcdf netcdf # Output trajectory, in binary netCDF format.
To start our analysis, we are going to put this new generated trajectory in the Trajectory File input box, and the associated topology (the same file used for the molecular dynamics simulation in case of keeping all the molecules or the new topology in case of having removed water molecules) in the Topology File input box.

Once we have our trajectory uploaded, we can begin with the analysis. NAFlex offers a set of basic analysis that can be divided in 3 different sections:
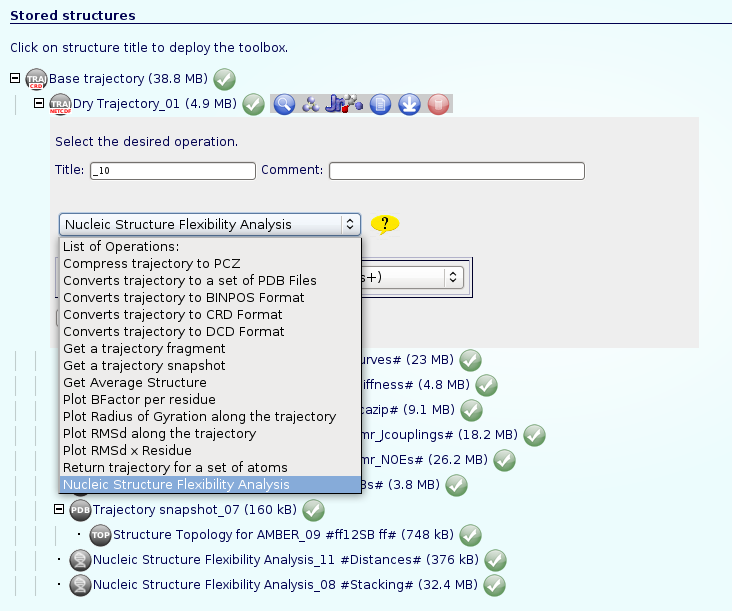
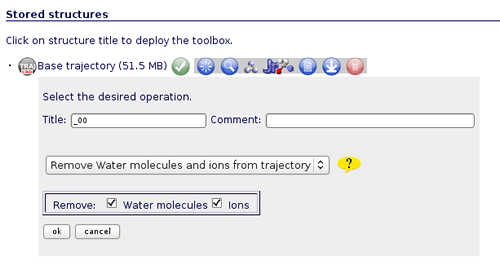
The most usual and important operation to do first of all is stripping the solvent (if it is not already done), usually toghether with the counterions, because the majority of the analysis work with the "dry" trajectory.
To do that, click on the operation called Remove water molecules and ions from trajectory. We can choose whether we would like to remove only water molecules, only counterions or both. It is recommended to remove both, obtaining the trajectory (and the correct topology associated) for just the structure of interest.
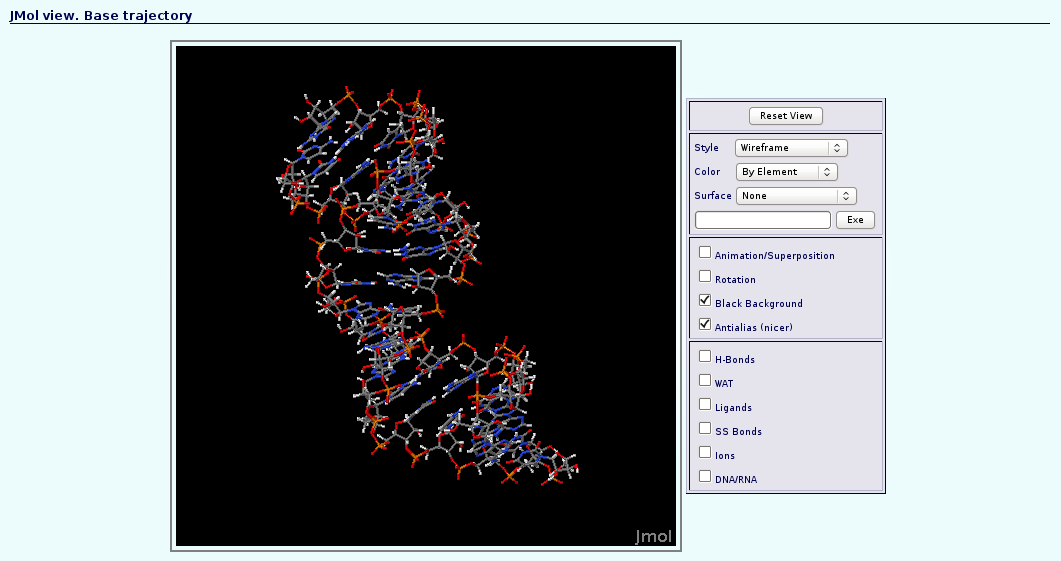
The first noticeable thing once we have the "dry" trajectory, is that it is really smaller in size than the solvated one. That allows the user to animate the trajectory with JMol applet. If we try to visualize the original solvated structure, NAFlex will show a warning message because the trajectory is too large, recommending to remove solvent. So, clicking at the corresponding JMol icon ![]() , a new window with the JMol interactive visualizer will be opened. It is a good first aproximation to take a look at the dynamics of the structure, before obtaining analysis data.
, a new window with the JMol interactive visualizer will be opened. It is a good first aproximation to take a look at the dynamics of the structure, before obtaining analysis data.
Now we can begin with the basic data analysis. From the list, we can choose basic analysis as Root Mean Square deviation and Radius of Gyration, showing us the deviation of the structure during the dynamics, whether it maintains its conformation during the simulation or it suffers some distortion.
Each of the analysis plot can be visualized clicking at the correspondent icon ![]() .
.
If we are interested in more complex analysis about the flexibility of our nucleic acid structure, NAFlex implements a complete set of Nucleic Acid Flexibility analysis. An independent help section is dedicated specifically to these analysis: NAFlex Analysis Tutorial.