NAFlex ABC Setup Pipeline Tutorial
NAFlex provides an ABC Setup pipeline to prepare a molecule to run a molecular dynamics simulation, following the steps used in ABC project: ABC NAFlex pipeline
Tutorial Steps
- ABC login
- Starting Project
- Checking the Structure
- Structure Setup
- Waiting Results
- Getting Results
The first thing to do is login to the ABC NAFlex pipeline server as an ABC user.
The first thing to do is choose between working as an anonymous abc user or alternatively as a registered user. We strongly recommend working as a registered user, as it has some important advantages.
Anonymous (abc) user's projects are completely removed once the user is disconnected and also when session expires (after some minutes of inactivity), and therefore working as anonymous (abc) user is only suited for a first impression of the server tool.
Registration process will just take a minute --> Registration.
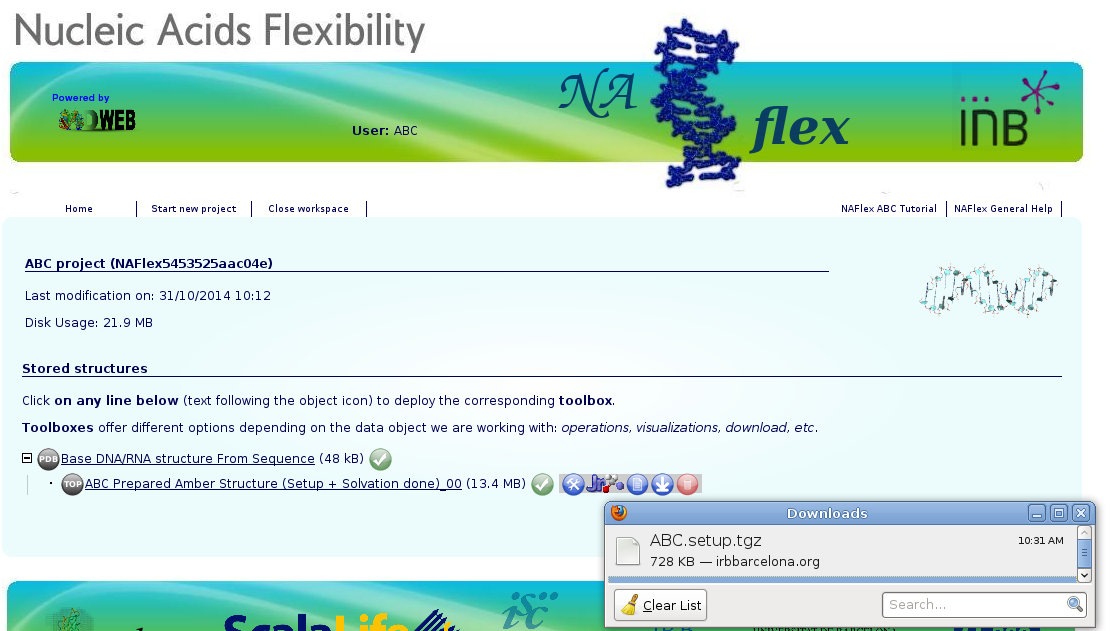
Once logged in, the ABC user workspace appears. In this workspace, all projects of the ABC user will be shown.
Now, we are ready to start our first ABC NAFlex project.

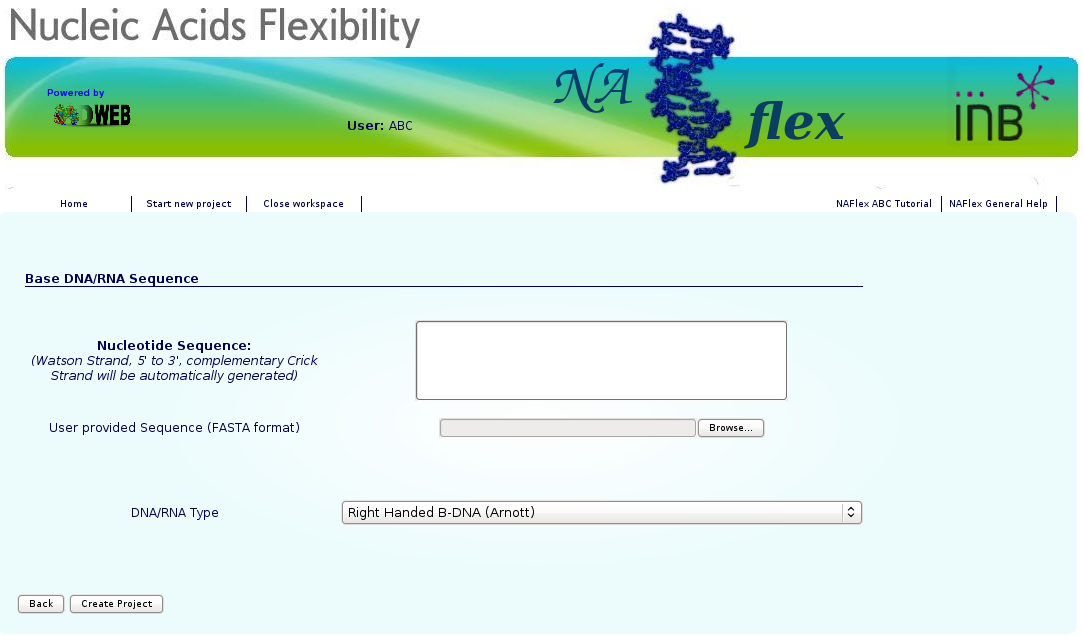
NAFlex user can choose between two different kind of inputs, DNA/RNA simulation from sequence or Simulation from PDB structure.
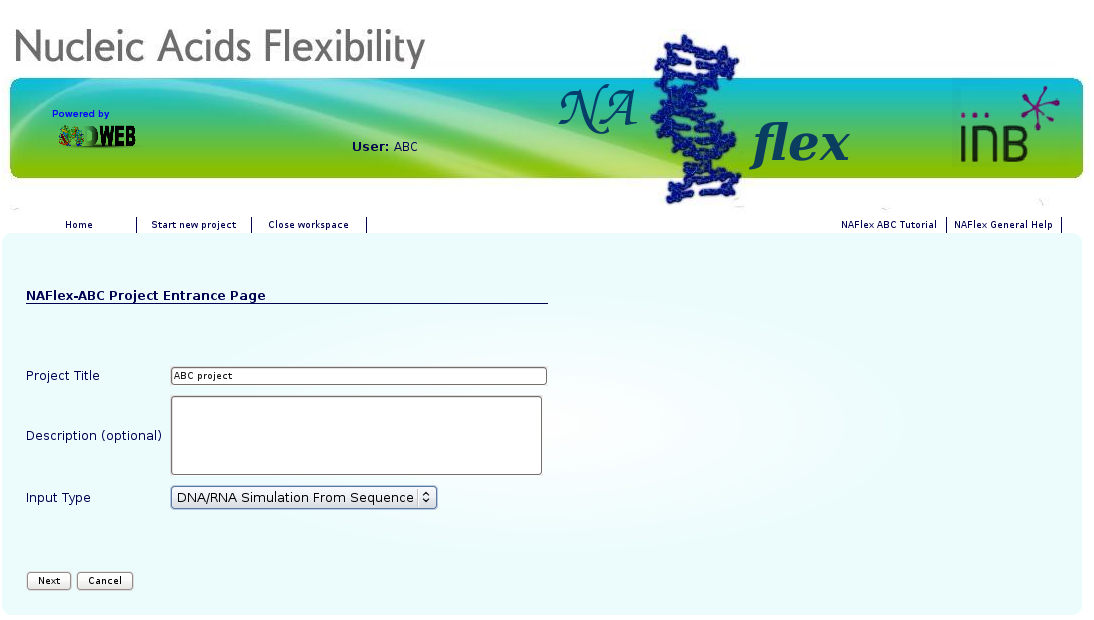
You will be asked for just three easy inputs: a project title, an optional description of the project, and an input nucleotide sequence, either directly typed or from a FASTA file. Alternatively, a PDB file can also be used.
Just after creating the project, you will be redirected to a Checking page. In this page, NAFlex will show a list of possible problems found in the input structure that can affect to a future Molecular Dynamics Simulation. User can interactively inspect these problems thanks to a JMol applet coupled to each of the problems, helping to locate them on the real tridimensional structure.
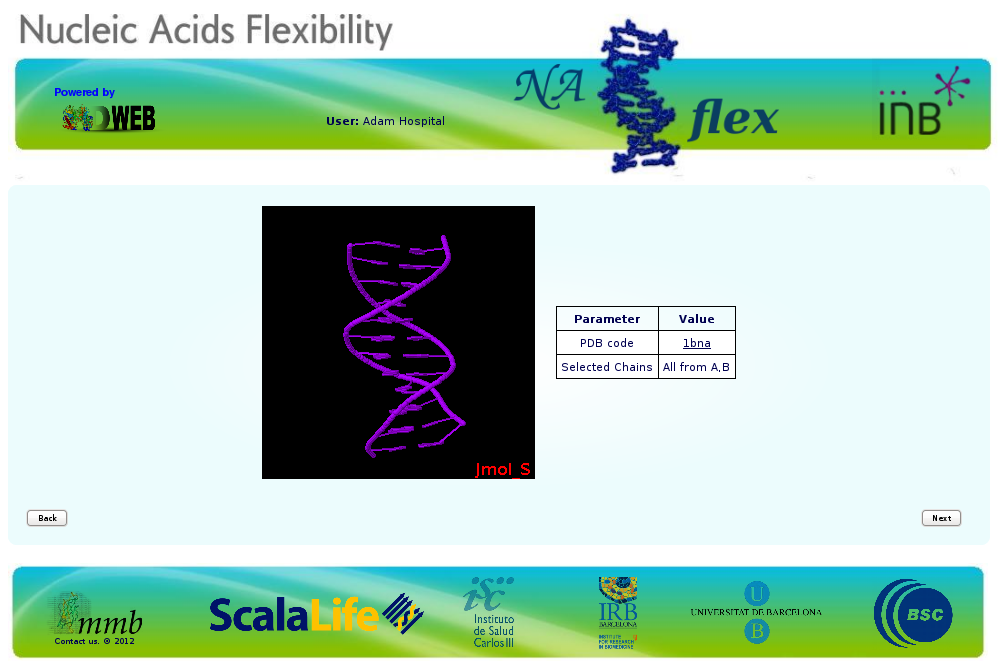
In this particular case, we are trying to work with the well-known B-DNA dodecamer d(CpGpCpGpApApTpTpCpGpCpG) (pdb code 1bna). At the right part of the screen, you will find all the possible problems identified, whereas at the left part of the screen the JMol applet with the structure loaded will be shown.

When clicking the next button, an intermediate information page will appear, listing the most important information chosen in the previous Checking page, and showing the final structure that will be used in the ABC pipeline.
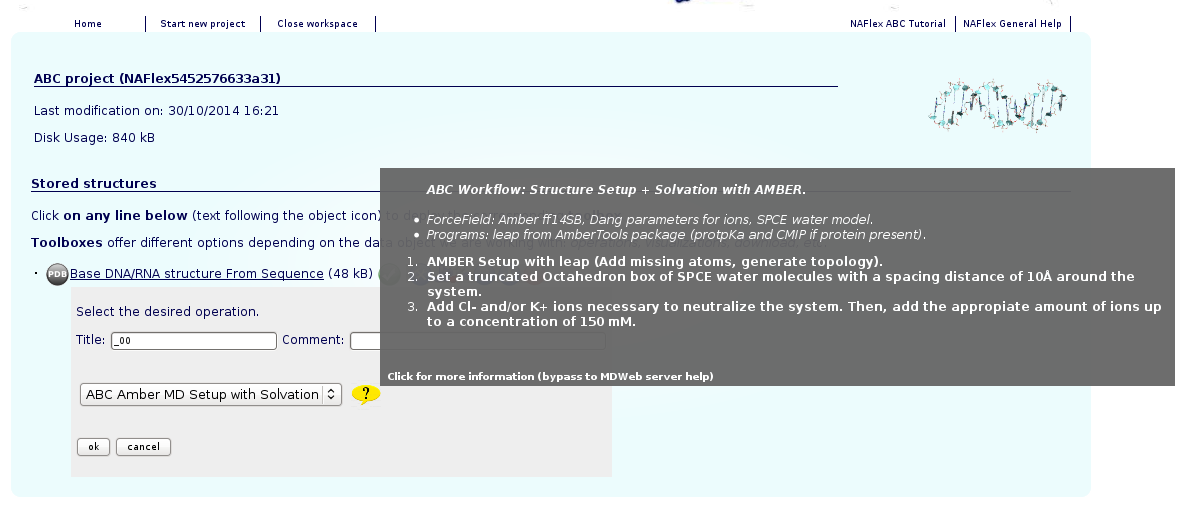
At this point, the structure is already loaded into NAFlex workspace. The next step is run the ABC Setup workflow. For that, the ABC Setup icon ![]() should be clicked, and the only operation available (ABC Amber MD Setup with Solvation) should be chosen. This pipeline will run the ABC setup pipeline, consisting on a set of processes performed with leap program from AMBER package, to obtain a solvated system with a determined ionic concentration.
should be clicked, and the only operation available (ABC Amber MD Setup with Solvation) should be chosen. This pipeline will run the ABC setup pipeline, consisting on a set of processes performed with leap program from AMBER package, to obtain a solvated system with a determined ionic concentration.
For a more extended information about the steps done by the workflow, see the corresponding NAFlex help section (ABC Amber Setup with Solvation).
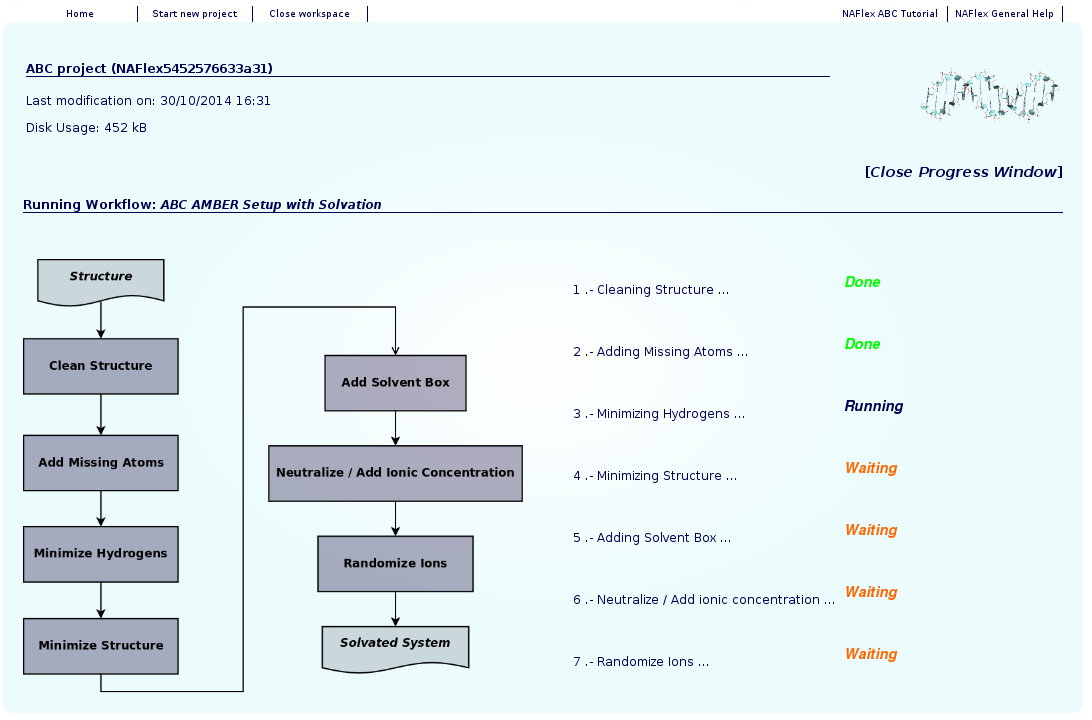
Once the workflow is launched, we just have to wait for the results. NAFlex will be automatically reloaded from time to time after checking if the process has already finished. Meanwhile, you can follow the progress of it clicking at Progress Info whenever you want.

When clicking at Progress Info, a new page will be opened showing the progress of the workflow step by step, accompanied by a figure of the whole workflow. This way, you will be able to know at any moment which service is being executed, how many steps have been already computed, and how many steps left.
Once the workflow has finished, the data generated (pdb structure, topology and coordinates) can be downloaded, just clicking at the download icon ![]() .
.