Following essential dynamics algorithm (ED), the orthogonal movements describing the variance of a system is obtained by diagonalization of the covariance matrix. The trajectory needed can be obtained from NMA-based MC, DMD or BD simulations. Note that for a complete space definition NMA eigenvectors and ED (NMA-based) eigenvectors are equivalent, but if a reduced set of NMA-eigenvectors/eigenvalues is used divergence between NMA and ED/NMA results is possible.
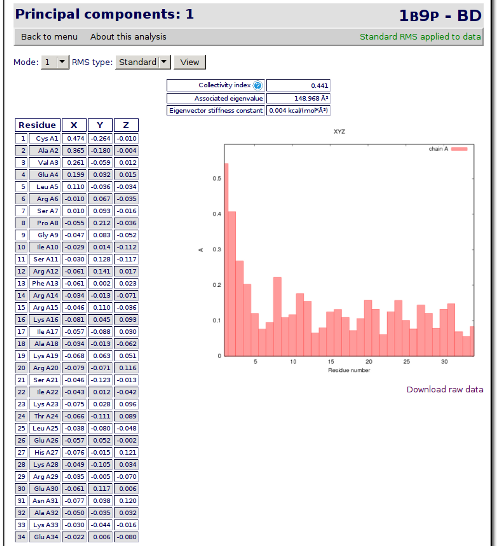
The result of the analysis is the generation of a set of eigenvectors (the modes or the principal components), which describe the nature of the deformation movements of the protein and a set of eigenvalues, which indicate the stiffness associated to every mode. By default the eigenvalues appear in distance² units, but can be transformed in energy units using harmonic approximation.
The eigenvectors appear ranked after a principal component analysis, the first one is that explaining the largest part of the variance (as indicated by the associated eigenvalue). Since the eigenvectors represent a full-basis set the original Cartesian trajectory can be always projected into the eigenvectors space, without lost of information. Furthermore, if a restricted set of eigenvectors is used information is lost, but the level of error introduced in the simplification is always on user-control by considering the annihilated variance (the residual value between the variance explained by the set of the eigenvectors considered and the total variance).
Inspection of the atomic components of the most important eigenvalues provided by this option helps to determine the contribution of different residues to the key essential deformations of the protein.
