Supplementary Tables
- Base Pair Step Average Helical Parameters
- Intra-Base Pair Average Helical Parameters
- Base Pair Axis Average Helical Parameters
- Base Pair Step Average Stiffness Constants
Average Helical Parameters and standard deviations for the unique base pair steps
(Supplementary table S3)
- Amount of data(N), average values and standard deviation of the six intra-strand base pair parameters for the 10 unique bps.
- For each step, the naked-DNA structures (first row), all the PDB structures (second row), and results from MD simulations with parmbsc0 (third row) are shown.
- Values for simulations were obtained from time averages computed for individual steps in each sequence.
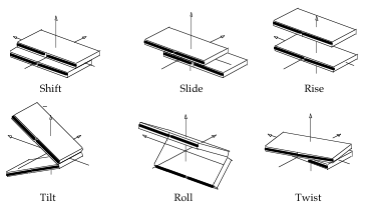
- Rotational parameters are in degrees (Twist, Tilt, Roll), and translational ones (Shift, Slide, Rise) in Å.
- Complementary steps (e.g. AG and CT) have the same average except for a change in sign of Shift and Tilt, and the same standard deviation.
| Step | Amount of data | Twist | Tilt | Roll | Shift | Slide | Rise |
| AA | 60 | 35.6±4.3 | -1.4±2.7 | 1.0±3.8 | 0.0±0.3 | -0.2±0.4 | 3.3±0.1 |
| AA | 850 | 33.4±5.9 | -1.2±3.0 | 2.5±7.1 | 0.1±0.5 | 0.0±0.6 | 3.3±0.2 |
| AA | 1M | 35.4±5.4 | -2.6±4.1 | 0.3±5.6 | -0.3±0.6 | -0.3±0.6 | 3.3±0.3 |
| TT | 60 | 35.6±4.3 | -1.4±2.7 | 1.0±3.8 | 0.0±0.3 | -0.2±0.4 | 3.3±0.1 |
| TT | 850 | 33.4±5.9 | -1.2±3.0 | 2.5±7.1 | 0.1±0.5 | 0.0±0.6 | 3.3±0.2 |
| TT | 1M | 35.4±5.4 | -2.6±4.1 | 0.3±5.6 | -0.3±0.6 | -0.3±0.6 | 3.3±0.3 |
| AC | 12 | 35.6±2.3 | -0.5±2.2 | 0.4±3.6 | -0.1±0.6 | -0.2±0.3 | 3.3±0.1 |
| AC | 670 | 31.5±3.9 | 0.7±3.4 | 2.0±4.1 | 0.2±0.6 | -0.6±0.4 | 3.2±0.2 |
| AC | 1M | 32.0±5.1 | -0.7±4.0 | -0.6±5.9 | 0.1±0.7 | -0.6±0.5 | 3.3±0.3 |
| GT | 12 | 35.6±2.3 | -0.5±2.2 | 0.4±3.6 | -0.1±0.6 | -0.2±0.3 | 3.3±0.1 |
| GT | 670 | 31.5±3.9 | 0.7±3.4 | 2.0±4.1 | 0.2±0.6 | -0.6±0.4 | 3.2±0.2 |
| GT | 1M | 32.0±5.1 | -0.7±4.0 | -0.6±5.9 | 0.1±0.7 | -0.6±0.5 | 3.3±0.3 |
| AG | 17 | 29.7±5.9 | -0.4±4.0 | 5.7±4.9 | 0.4±0.4 | 0.3±0.8 | 3.4±0.3 |
| AG | 700 | 32.4±4.7 | -1.1±3.6 | 3.2±5.7 | 0.1±0.7 | -0.2±0.3 | 3.3±0.3 |
| AG | 1M | 33.5±6.0 | -2.5±4.2 | 3.1±5.9 | -0.4±0.7 | -0.6±0.7 | 3.4±0.3 |
| CT | 17 | 29.7±5.9 | -0.4±4.0 | 5.7±4.9 | 0.4±0.4 | 0.3±0.8 | 3.4±0.3 |
| CT | 700 | 32.4±4.7 | -1.1±3.6 | 3.2±5.7 | 0.1±0.7 | -0.2±0.3 | 3.3±0.3 |
| CT | 1M | 33.5±6.0 | -2.5±4.2 | 3.1±5.9 | -0.4±0.7 | -0.6±0.7 | 3.4±0.3 |
| AT | 62 | 32.0±3.6 | 0.3±2.2 | -1.2±3.1 | 0.1±0.4 | -0.6±0.3 | 3.3±0.2 |
| AT | 890 | 30.0±4.7 | 0.2±2.9 | 1.6±5.3 | 0.0±0.5 | -0.6±0.4 | 3.2±0.3 |
| AT | 1M | 30.4±4.2 | 0.0±3.8 | -0.5±5.1 | 0.0±0.7 | -0.8±0.4 | 3.3±0.3 |
| CA | 30 | 41.9±8.7 | 0.7±2.7 | 0.7±7.0 | 0.1±0.2 | 1.6±1.0 | 3.3±0.2 |
| CA | 730 | 35.9±5.5 | 0.3±3.9 | 4.6±6.8 | -0.1±0.6 | 0.6±1.0 | 3.4±0.3 |
| CA | 1M | 29.6±8.3 | 0.2±4.8 | 10.3±6.3 | -0.2±0.7 | -0.2±0.6 | 3.1±0.4 |
| TG | 30 | 41.9±8.7 | 0.7±2.7 | 0.7±7.0 | 0.1±0.2 | 1.6±1.0 | 3.3±0.2 |
| TG | 730 | 35.9±5.5 | 0.3±3.9 | 4.6±6.8 | -0.1±0.6 | 0.6±1.0 | 3.4±0.3 |
| TG | 1M | 29.6±8.3 | 0.2±4.8 | 10.3±6.3 | -0.2±0.7 | -0.2±0.6 | 3.1±0.4 |
| CC | 36 | 33.8±5.6 | 1.9±3.4 | 4.3±4.6 | 0.0±0.5 | 0.7±1.0 | 3.4±0.3 |
| CC | 560 | 33.7±4.6 | 0.9±4.7 | 4.3±5.5 | 0.1±0.7 | -0.2±0.8 | 3.4±0.3 |
| CC | 1M | 32.7±6.6 | 0.0±4.4 | 4.9±5.7 | 0.2±0.7 | -0.7±0.9 | 3.5±0.3 |
| GG | 36 | 33.8±5.6 | 1.9±3.4 | 4.3±4.6 | 0.0±0.5 | 0.7±1.0 | 3.4±0.3 |
| GG | 560 | 33.7±4.6 | 0.9±4.7 | 4.3±5.5 | 0.1±0.7 | -0.2±0.8 | 3.4±0.3 |
| GG | 1M | 32.7±6.6 | 0.0±4.4 | 4.9±5.7 | 0.2±0.7 | -0.7±0.9 | 3.5±0.3 |
| CG | 205 | 33.0±5.1 | 0.3±4.5 | 3.4±7.7 | 0.1±0.5 | 0.5±0.6 | 3.3±0.3 |
| CG | 860 | 32.5±5.7 | -0.2±4.2 | 4.6±5.9 | 0.0±0.7 | 0.4±0.8 | 3.3±0.3 |
| CG | 1M | 29.8±8.8 | 0.0±5.2 | 9.3±6.0 | 0.0±0.7 | 0.0±0.5 | 3.1±0.4 |
| GA | 35 | 38.9±3.9 | -0.8±3.3 | 2.3±3.8 | 0.0±0.4 | 0.0±0.5 | 3.4±0.2 |
| GA | 805 | 35.4±4.7 | -1.4±3.6 | 2.3±5.5 | -0.2±0.6 | -0.1±0.6 | 3.3±0.2 |
| GA | 1M | 36.6±6.2 | -1.6±4.5 | 1.6±5.9 | -0.4±0.7 | -0.3±0.7 | 3.4±0.3 |
| TC | 35 | 38.9±3.9 | -0.8±3.3 | 2.3±3.8 | 0.0±0.4 | 0.0±0.5 | 3.4±0.2 |
| TC | 805 | 35.4±4.7 | -1.4±3.6 | 2.3±5.5 | -0.2±0.6 | -0.1±0.6 | 3.3±0.2 |
| TC | 1M | 36.6±6.2 | -1.6±4.5 | 1.6±5.9 | -0.4±0.7 | -0.3±0.7 | 3.4±0.3 |
| GC | 125 | 39.7±4.4 | -0.5±3.7 | -5.2±6.0 | -0.2±0.8 | 0.3±0.4 | 3.5±0.2 |
| GC | 940 | 35.0±5.2 | -0.2±3.7 | 0.6±5.7 | 0.0±0.7 | -0.1±0.6 | 3.3±0.3 |
| GC | 1M | 35.7±5.1 | 0.0±4.0 | -1.3±5.8 | 0.0±0.7 | -0.4±0.6 | 3.5±0.3 |
| TA | 19 | 41.7±6.3 | -0.1±3.5 | 1.4±5.0 | -0.2±0.4 | 0.7±0.8 | 3.4±0.2 |
| TA | 680 | 35.7±7.5 | 0.1±3.6 | 3.2±7.2 | 0.0±0.6 | 0.3±1.0 | 3.4±0.4 |
| TA | 25000 | 28.9±8.4 | 0.0±5.1 | 10.0±7.4 | 0.0±0.9 | -0.2±0.8 | 3.2±0.4 |
Units:
Translational Inter-BP Step Helical Parameters Shift/Slide/Rise in Å, Rotational Inter-BP Helical Parameters Tilt/Roll/Twist in degrees
Average Intra-Base Pair Helical Parameters and standard deviations for the AT, TA, CG, GC base pairs
(Supplementary table S3)
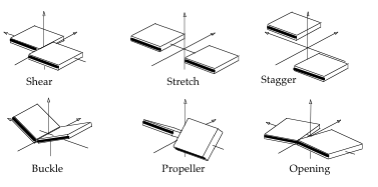
| Base Pair | Shear | Stretch | Stagger | Buckle | Propeller | Opening |
| AT | 0.10±0.29 | 0.03±0.12 | 0.07±0.42 | 3.7±11.7 | -12.9±9.0 | 3.1±5.5 |
| GC | 0.02±0.31 | 0.03±0.11 | 0.10±0.39 | 2.8±12.3 | -9.1±9.2 | 1.1±3.3 |
| TA | 0.10±0.29 | 0.03±0.12 | 0.07±0.42 | 3.7±11.7 | -12.9±9.0 | 3.1±5.5 |
| CG | 0.02±0.31 | 0.03±0.11 | 0.10±0.39 | 2.8±12.3 | -9.1±9.2 | 1.1±3.3 |
Units: Translational Intra-BP Helical Parameters Shear/Stretch/Stagger in Å, Rotational Intra-BP Helical Parameters Buckle/Propeller/Opening in degrees.
Average Base Pair Axis Helical Parameters and standard deviations for the AT, TA, CG, GC base pairs
(Supplementary table S3)
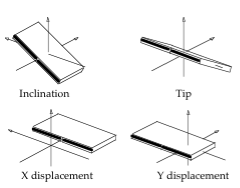
| Base Pair | Xdisp | Ydisp | Inclination | Tip |
| AT | -1.45±0.82 | 0.10±0.53 | 6.7±5.5 | 0.2±5.0 |
| GC | -1.43±0.96 | 0.00±0.56 | 6.9±5.2 | 0.5±4.9 |
| TA | -1.45±0.82 | 0.10±0.53 | 6.7±5.5 | 0.2±5.0 |
| CG | -1.43±0.96 | 0.00±0.56 | 6.9±5.2 | 0.5±4.9 |
Units: Translational BP Axis Helical Parameters Xdisp/Ydisp in Å, Rotational BP Axis Helical Parameters Inclination/Tip in degrees.
Average Stiffness Constants for the unique base pair steps

| Base Pair | Shift | Slide | Rise | Tilt | Roll | Twist |
| AATT | ||||||
| 1.72017 | 0.19796 | 0.32533 | -0.01249 | 0.00576 | 0.05913 | |
| 0.19797 | 2.12618 | 0.75074 | -0.00581 | -0.05309 | -0.10162 | |
| 0.32534 | 0.75074 | 7.64359 | -0.18348 | -0.04547 | -0.14850 | |
| -0.01249 | -0.00581 | -0.18349 | 0.03738 | 0.00211 | 0.00597 | |
| 0.00576 | -0.05309 | -0.04547 | 0.00211 | 0.01961 | 0.00742 | |
| 0.05913 | -0.10162 | -0.14850 | 0.00597 | 0.00742 | 0.02761 | |
| -0.18750 | -0.41750 | 3.32750 | -2.01750 | 2.73750 | 33.57250 | |
| ACGT | ||||||
| 1.28288 | 0.13127 | 0.29502 | -0.02780 | 0.00302 | 0.03646 | |
| 0.13127 | 2.97699 | 2.10518 | -0.02280 | 0.03038 | -0.10881 | |
| 0.29502 | 2.10518 | 8.83137 | 0.04907 | 0.10478 | -0.14741 | |
| -0.02780 | -0.02280 | 0.04907 | 0.03776 | 0.00378 | 0.00418 | |
| 0.00302 | 0.03038 | 0.10479 | 0.00378 | 0.02306 | 0.00708 | |
| 0.03646 | -0.10881 | -0.14742 | 0.00418 | 0.00708 | 0.03551 | |
| 0.05000 | -0.73571 | 3.37143 | -1.06286 | 0.00857 | 33.38429 | |
| AGCT | ||||||
| 1.39990 | 0.27887 | 0.27572 | -0.03917 | 0.02080 | 0.07408 | |
| 0.27887 | 1.78493 | 0.99427 | -0.00395 | -0.01181 | -0.06894 | |
| 0.27572 | 0.99427 | 7.04130 | -0.15259 | -0.02113 | -0.14069 | |
| -0.03917 | -0.00395 | -0.15259 | 0.03699 | 0.00410 | 0.00630 | |
| 0.02080 | -0.01181 | -0.02113 | 0.00410 | 0.01912 | 0.00500 | |
| 0.07408 | -0.06894 | -0.14069 | 0.00630 | 0.00500 | 0.02797 | |
| -0.32750 | -0.65000 | 3.38750 | -2.70500 | 2.83250 | 32.05750 | |
| ATAT | ||||||
| 1.04661 | 0.00000 | 0.00000 | 0.03066 | 0.00000 | 0.00000 | |
| 0.00000 | 3.76524 | 2.16877 | 0.00000 | -0.03029 | -0.04352 | |
| 0.00000 | 2.16877 | 9.33735 | 0.00000 | 0.06011 | -0.09916 | |
| 0.03066 | 0.00000 | 0.00000 | 0.03494 | 0.00000 | 0.00000 | |
| 0.00000 | -0.03029 | 0.06011 | 0.00000 | 0.02157 | 0.00877 | |
| 0.00000 | -0.04352 | -0.09916 | 0.00000 | 0.00877 | 0.03122 | |
| 0.00000 | -0.77333 | 3.14333 | 0.00000 | 1.47000 | 28.59333 | |
| CATG | ||||||
| 1.05328 | 0.07552 | 0.23196 | -0.03375 | 0.00158 | -0.01277 | |
| 0.07552 | 1.79677 | 0.52999 | -0.00472 | 0.02114 | -0.03759 | |
| 0.23197 | 0.53000 | 6.30474 | 0.01221 | -0.07708 | -0.14400 | |
| -0.03375 | -0.00472 | 0.01221 | 0.02498 | -0.00127 | 0.00137 | |
| 0.00158 | 0.02114 | -0.07708 | -0.00127 | 0.01647 | 0.00541 | |
| -0.01277 | -0.03759 | -0.14399 | 0.00137 | 0.00541 | 0.01534 | |
| -0.32600 | -0.22200 | 3.20800 | 0.42800 | 9.24600 | 28.43600 | |
| CCGG | ||||||
| 1.43205 | -0.29768 | -0.35747 | -0.08468 | -0.01448 | -0.03982 | |
| -0.29768 | 1.57310 | 1.18241 | -0.00677 | 0.01068 | -0.09303 | |
| -0.35748 | 1.18243 | 7.85985 | 0.25771 | 0.00156 | -0.11971 | |
| -0.08468 | -0.00677 | 0.25771 | 0.04202 | 0.00134 | 0.00022 | |
| -0.01448 | 0.01068 | 0.00156 | 0.00134 | 0.02012 | 0.00493 | |
| -0.03982 | -0.09303 | -0.11970 | 0.00022 | 0.00493 | 0.02602 | |
| 0.11125 | -0.76000 | 3.55625 | -0.43625 | 4.58000 | 34.18625 | |
| CGCG | ||||||
| 1.05459 | 0.00000 | 0.00000 | -0.07841 | 0.00000 | 0.00000 | |
| 0.00000 | 1.91048 | 0.56341 | 0.00000 | 0.02388 | -0.05037 | |
| 0.00000 | 0.56342 | 6.11191 | 0.00000 | -0.04625 | -0.14832 | |
| -0.07841 | 0.00000 | 0.00000 | 0.02587 | 0.00000 | 0.00000 | |
| 0.00000 | 0.02388 | -0.04625 | 0.00000 | 0.01562 | 0.00319 | |
| 0.00000 | -0.05037 | -0.14832 | 0.00000 | 0.00319 | 0.01400 | |
| 0.00000 | -0.07250 | 3.25875 | 0.00000 | 6.45000 | 31.88875 | |
| CTAG | ||||||
| 1.39990 | -0.27887 | -0.27572 | -0.03917 | -0.02080 | -0.07408 | |
| -0.27887 | 1.78493 | 0.99427 | 0.00395 | -0.01181 | -0.06894 | |
| -0.27572 | 0.99427 | 7.04130 | 0.15259 | -0.02113 | -0.14069 | |
| -0.03917 | 0.00395 | 0.15259 | 0.03699 | -0.00410 | -0.00630 | |
| -0.02080 | -0.01181 | -0.02113 | -0.00410 | 0.01912 | 0.00500 | |
| -0.07408 | -0.06894 | -0.14069 | -0.00630 | 0.00500 | 0.02797 | |
| 0.32750 | -0.65000 | 3.38750 | 2.70500 | 2.83250 | 32.05750 | |
| GATC | ||||||
| 1.31766 | 0.29517 | 0.40655 | -0.04665 | 0.00977 | 0.01782 | |
| 0.29517 | 1.87898 | 1.00952 | 0.00116 | -0.01328 | -0.09736 | |
| 0.40656 | 1.00952 | 8.48201 | -0.25663 | -0.01013 | -0.12376 | |
| -0.04665 | 0.00116 | -0.25663 | 0.03758 | -0.00252 | 0.00117 | |
| 0.00977 | -0.01328 | -0.01012 | -0.00252 | 0.02025 | 0.00861 | |
| 0.01782 | -0.09736 | -0.12376 | 0.00117 | 0.00861 | 0.02441 | |
| -0.21400 | -0.29000 | 3.37200 | -0.49800 | 2.13600 | 36.47000 | |
| GCGC | ||||||
| 1.17900 | 0.00000 | 0.00000 | -0.08357 | 0.00000 | 0.00000 | |
| 0.00000 | 2.58821 | 2.05067 | 0.00000 | 0.08753 | -0.07364 | |
| 0.00000 | 2.05063 | 9.46559 | 0.00000 | 0.15720 | -0.18431 | |
| -0.08357 | 0.00000 | 0.00000 | 0.03618 | 0.00000 | 0.00000 | |
| 0.00000 | 0.08753 | 0.15720 | 0.00000 | 0.02569 | 0.00442 | |
| 0.00000 | -0.07364 | -0.18431 | 0.00000 | 0.00442 | 0.02238 | |
| 0.00000 | -0.28800 | 3.41000 | 0.00000 | -1.07600 | 36.07200 | |
| GGCC | ||||||
| 1.43205 | 0.29768 | 0.35747 | -0.08468 | 0.01448 | 0.03982 | |
| 0.29768 | 1.57310 | 1.18241 | 0.00677 | 0.01068 | -0.09303 | |
| 0.35748 | 1.18243 | 7.85985 | -0.25771 | 0.00156 | -0.11971 | |
| -0.08468 | 0.00677 | -0.25771 | 0.04202 | -0.00134 | -0.00022 | |
| 0.01448 | 0.01068 | 0.00156 | -0.00134 | 0.02012 | 0.00493 | |
| 0.03982 | -0.09303 | -0.11970 | -0.00022 | 0.00493 | 0.02602 | |
| -0.11125 | -0.76000 | 3.55625 | 0.43625 | 4.58000 | 34.18625 | |
| GTAC | ||||||
| 1.28288 | -0.13127 | -0.29502 | -0.02780 | -0.00302 | -0.03646 | |
| -0.13127 | 2.97699 | 2.10518 | 0.02280 | 0.03038 | -0.10881 | |
| -0.29502 | 2.10518 | 8.83137 | -0.04907 | 0.10478 | -0.14741 | |
| -0.02780 | 0.02280 | -0.04907 | 0.03776 | -0.00378 | -0.00418 | |
| -0.00302 | 0.03038 | 0.10479 | -0.00378 | 0.02306 | 0.00708 | |
| -0.03646 | -0.10881 | -0.14742 | -0.00418 | 0.00708 | 0.03551 | |
| -0.05000 | -0.73571 | 3.37143 | 1.06286 | 0.00857 | 33.38429 | |
| TATA | ||||||
| 0.64315 | 0.00000 | 0.00000 | -0.01544 | 0.00000 | 0.00000 | |
| 0.00000 | 1.24864 | 0.53653 | 0.00000 | 0.02038 | -0.04224 | |
| 0.00000 | 0.53653 | 6.07971 | 0.00000 | -0.07916 | -0.14834 | |
| -0.01544 | 0.00000 | 0.00000 | 0.01900 | 0.00000 | 0.00000 | |
| 0.00000 | 0.02038 | -0.07916 | 0.00000 | 0.01464 | 0.00831 | |
| 0.00000 | -0.04224 | -0.14835 | 0.00000 | 0.00831 | 0.01759 | |
| 0.00000 | -0.14667 | 3.26667 | 0.00000 | 8.31667 | 32.66000 | |
| TCGA | ||||||
| 1.31766 | -0.29517 | -0.40655 | -0.04665 | -0.00977 | -0.01782 | |
| -0.29517 | 1.87898 | 1.00952 | -0.00116 | -0.01328 | -0.09736 | |
| -0.40656 | 1.00952 | 8.48201 | 0.25663 | -0.01013 | -0.12376 | |
| -0.04665 | -0.00116 | 0.25663 | 0.03758 | 0.00252 | -0.00117 | |
| -0.00977 | -0.01328 | -0.01012 | 0.00252 | 0.02025 | 0.00861 | |
| -0.01782 | -0.09736 | -0.12376 | -0.00117 | 0.00861 | 0.02441 | |
| 0.21400 | -0.29000 | 3.37200 | 0.49800 | 2.13600 | 36.47000 | |
| TGCA | ||||||
| 1.05328 | -0.07552 | -0.23196 | -0.03375 | -0.00158 | 0.01277 | |
| -0.07552 | 1.79677 | 0.52999 | 0.00472 | 0.02114 | -0.03759 | |
| -0.23197 | 0.53000 | 6.30474 | -0.01221 | -0.07708 | -0.14400 | |
| -0.03375 | 0.00472 | -0.01221 | 0.02498 | 0.00127 | -0.00137 | |
| -0.00158 | 0.02114 | -0.07708 | 0.00127 | 0.01647 | 0.00541 | |
| 0.01277 | -0.03759 | -0.14399 | -0.00137 | 0.00541 | 0.01534 | |
| 0.32600 | -0.22200 | 3.20800 | -0.42800 | 9.24600 | 28.43600 | |
| TTAA | ||||||
| 1.72017 | -0.19796 | -0.32533 | -0.01249 | -0.00576 | -0.05913 | |
| -0.19797 | 2.12618 | 0.75074 | 0.00581 | -0.05309 | -0.10162 | |
| -0.32534 | 0.75074 | 7.64359 | 0.18348 | -0.04547 | -0.14850 | |
| -0.01249 | 0.00581 | 0.18349 | 0.03738 | -0.00211 | -0.00597 | |
| -0.00576 | -0.05309 | -0.04547 | -0.00211 | 0.01961 | 0.00742 | |
| -0.05913 | -0.10162 | -0.14850 | -0.00597 | 0.00742 | 0.02761 | |
| 0.18750 | -0.41750 | 3.32750 | 2.01750 | 2.73750 | 33.57250 |
Units:
Diagonal Shift/Slide/Rise in kcal/(mol*Ų), Diagonal Tilt/Roll/Twist in kcal/(mol*degree²)
Out of Diagonal: Shift/Slide/Rise in kcal/(mol*Å), Out of Diagonal Tilt/Roll/Twist in kcal/(mol*degree)