ParmBSC1 forcefield Nucleotide MD Simulations Database
----
ParmBSC1 Portfolio, a BigData approach to efficiently manage large nucleic acids simulation data.
Analyses
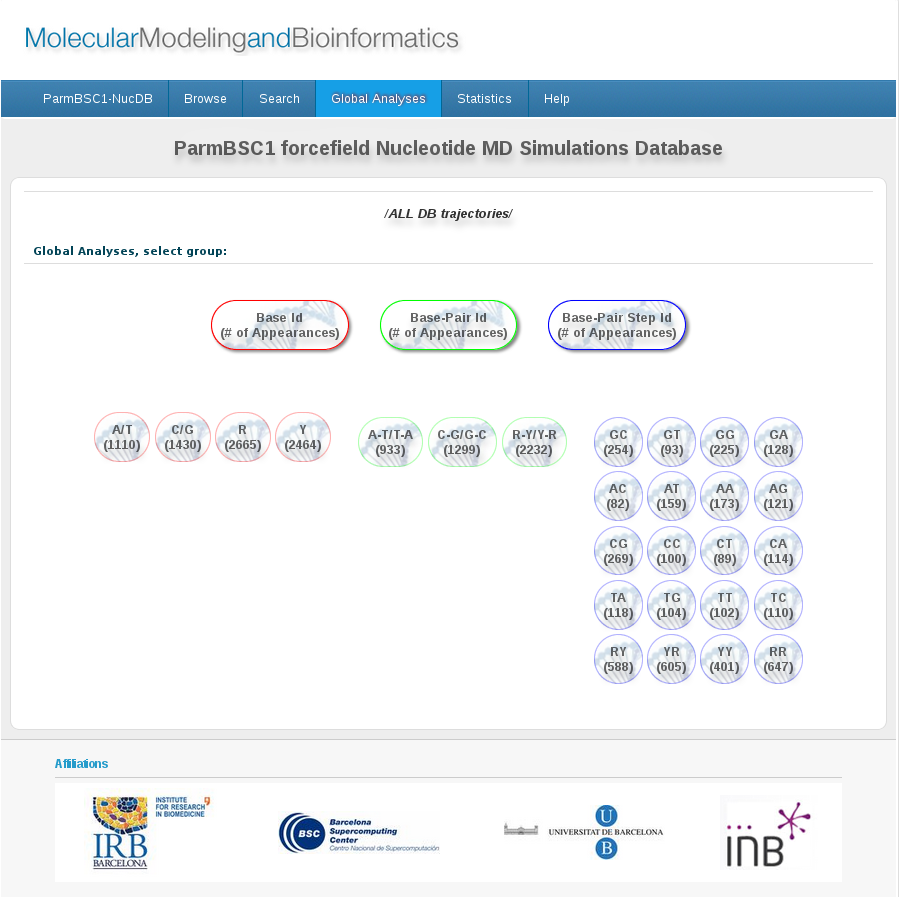
Global Analyses screen is designed to offer a very easy and intuitive way to access to base, base-pair and base-pair step analysis, either for the global set of trajectories stored in the database or for just a selection of interest (from the browse page).
Bullets with different colors represent: bases (red), base-pairs (green) and base-pair steps (blue) in one-letter code, defined below:
- A: Adenine
- C: Cytosine
- G: Guanine
- T: Thymine
- U: Uracil
- R: Purine (Adenine and Guanine)
- Y: Pyrimidine (Cytosine, Thymine, Uracil)
Each bullet show together with the base/base-pair/base-pair step code the number of appearances of it in the selected set of sequences.
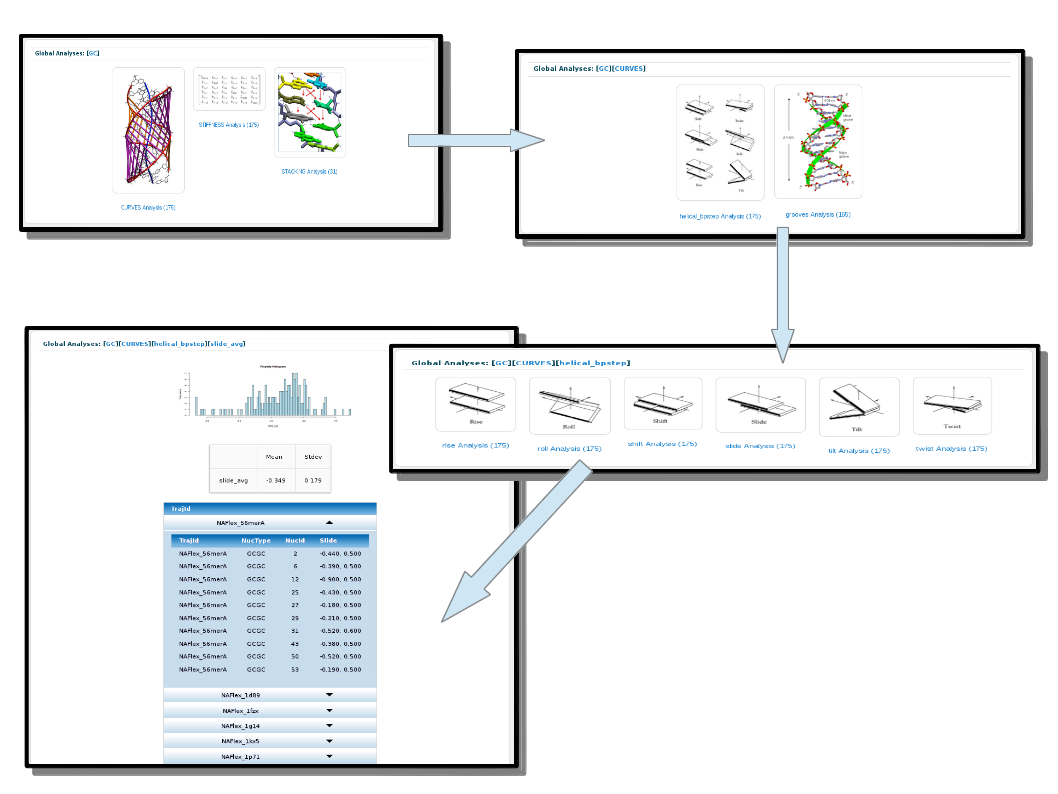
Clicking on one of the bullets will open a new section at the bottom of the page with all the analyses stored in the database for the particular code. As the analyses are defined in a hierarchical way, user must select the desired analysis at each step, until finding the final values. Final results are shown in different ways, depending on the analysis (histograms, average values, 2D plots, etc.).
As an example, if a user is interested in the Slide helical parameter values for the particular CG base-pair step, the steps to follow are described in the next figure: After clicking on the CG bullet, select Curves analysis, helical parameters, and finally Slide parameter.