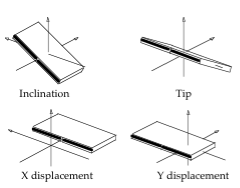
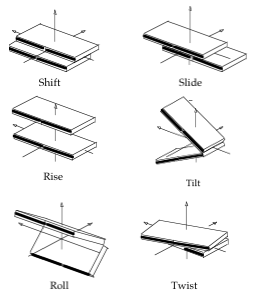
Entry: NAFlex_440d
Nucleic Sequence:
| Sequence | AGGGGCCCCT |
| Rev. Sequence | AGGGGCCCCT |
| Type | DNA |
| SubType | A |
| Chains | duplex |
| Pdb | 440D |
| Ligands | no |
MD Simulation >> (Click to expand/shrink)
Unified Molecular Modeling (UMM) Metadata
(Click to see full UMM)
(Click to see full UMM)
| AdditionalIons | No |
| AdditionalMolecules | No |
| AdditionalSolvent | No |
| Author | A.P. |
| Chains | duplex |
| Comments | /orozco/scratch/parmBSC1/Alberto2/440d |
| CounterIons | Na+ |
| Format | mdcrd |
| FrameStep | 1ps |
| Frames | 200000 |
| IonicConcentration | Electroneutrality |
| IonsParameters | Dang |
| Ligands | no |
| NucType | DNA |
| PDB | 440d |
| Parts | ions + DNA |
| RMSd_avg | 4.293 |
| RMSd_stdev | 0.541 |
| Rgyr_avg | 12.316 |
| Rgyr_stdev | 0.393 |
| SASA_avg | 3657.433 |
| SASA_stdev | 67.904 |
| SubType | A |
| Temperature | 300K |
| Topology | 440d.dry.top |
| Trajectory | 440d.trj |
| Water | TIP3P |
| _id | NAFlex_440d |
| altPDB | 440D 441D |
| authors | Ivan Ivani, Pablo D. Dans, Agnès Noy, Alberto Pérez, Ignacio Faustino, Adam Hospital, Jürgen Walther, Pau Andrio, Ramon Goñi, Alexandra Balaceanu, Guillem Portella, Federica Battistini, Josep Lluís Gelpí, Carlos González, Michele Vendruscolo, Charles A. Laughton, Sarah A. Harris, David A. Case and Modesto Orozco |
| dataset | Array |
| date | 0.00000000 1429920000 |
| description | DNA-A Duplex Naked ParmBSC1 TIP3P Electroneutral |
| forceField | parmBSC1 |
| ionsModel | - |
| moleculeType | Dna |
| rev_sequence | AGGGGCCCCT |
| saltConcentration | - |
| sequence | AGGGGCCCCT |
| soluteAtoms | 630 |
| soluteCharge | -18 |
| soluteResidues | 20 |
| solventAtoms | - |
| solventResidues | - |
| time | 200 |
| totalAtoms | 630 |
| totalCharge | -18 |
| totalIons | - |
| totalResidues | 20 |