Entry: NAFlex_Circular29_2_CG
Nucleic Acid Data:
| Sequence | TCTCTCTCTCTCTCTCACCACCGCGTACAAGAAAGTTTGTTCTTTCATTTCGCCATACAGCTGCCTCGCGCGTTTCGGTGATATCGGTGAAAACCCTTCCCGTTTCGCTCAAGTTAGTATAAAAAAGCTGAACGAGAAACGTAAAATGATATAAATATCAATATATTAAATTAGATTTTGCATAAAAAACAGACTACATAATACTGTAAAACACAACATATGCAGTCACTATGAATCAACTACTTAGATGGTATTAGTGACCTGTAACAGAGCTGAGGCATCTTTCTCAATGAAAGTGTCGGAACTCGGTACATAGGCTTCCGGCTGATAGTAGTCGTATACGACTACTATCAGCCGGAAGCCTATGTACCGAGTTCCGACACTTTCATTGAGAAAGATGCCTCAGCTCTGTTACAGGTCACTAATACCATCTAAGTAGTTGATTCATAGTGACTGCATATGTTGTGTTTTACAGTATTATGTAGTCTGTTTTTTATGCAAAATCTAATTTAATATATTGATATTTATATCATTTTACGTTTCTCGTTCAGCTTTTTTATACTAACTTGAGCGAAACGGGAAGGGTTTTCACCGATATCACCGAAACGCGCGAGGCAGCTGTATGGCGAAATGAAAGAACAAACTTTCTTGTACGCGGTGGTGAGAGAGAGAGAGAGA |
| Rev. Sequence | - |
| Type | DNA |
| SubType | B |
| Chains | duplex |
| Pdb | - |
| Ligands | No |
| Keywords | DNA-B Duplex Naked |
MD Simulation >> (Click to expand/shrink)
| Simulation Metadata | |
|---|---|
| Force Field | CGeNArate |
| Simulation Date | 2024-Jul-08 |
| Simulated Time | 1,000 ns |
| Time Step | 20 ps |
| Parts | DNA |
| Temperature | 298K |
| Water | No |
| Additional Solvent | No |
| Counter Ions | No |
| Ionic Concentration | No |
| Additional Ions | No |
| Additional Molecules | No |
| Ions Parameters | No |
Unified Molecular Modeling (UMM) Metadata
(Click to see full UMM)
(Click to see full UMM)
| Simulation Metadata (UMM) | |
|---|---|
| AdditionalIons | No |
| AdditionalMolecules | No |
| AdditionalSolvent | No |
| Author | D.F. |
| Chains | duplex |
| Comments | CGeNArate: A Sequence-Dependent Coarse-Grained Model of DNA for Accurate Atomistic MD Simulations of kb-long Duplexes, DOI: 10.1093/nar/gkae444 |
| CounterIons | No |
| Format | mdcrd |
| FrameStep | 20 ps |
| Frames | 50000 |
| IonicConcentration | No |
| IonsParameters | No |
| Ligands | No |
| NucType | DNA |
| PDB | NONE |
| Parts | DNA |
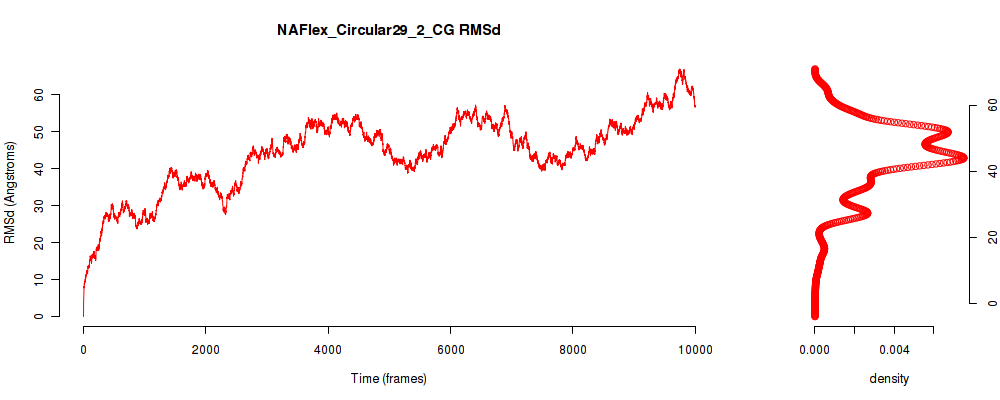
| RMSd_avg | 44.206 |
| RMSd_stdev | 10.257 |
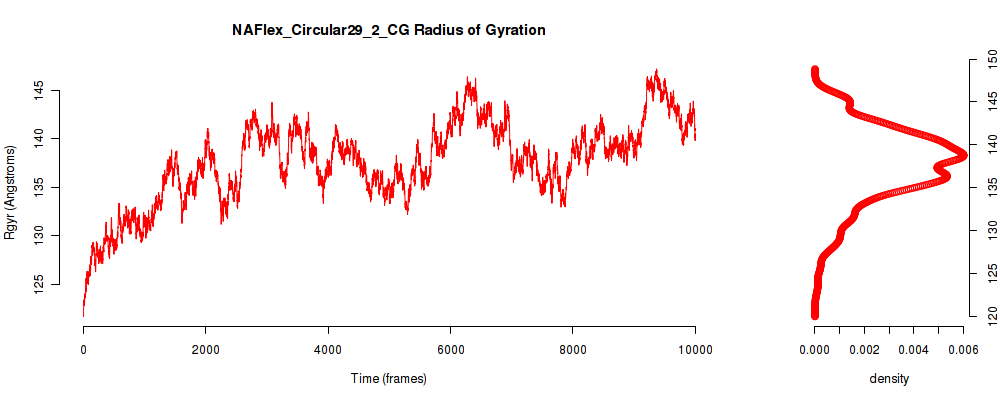
| Rgyr_avg | 137.620 |
| Rgyr_stdev | 4.428 |
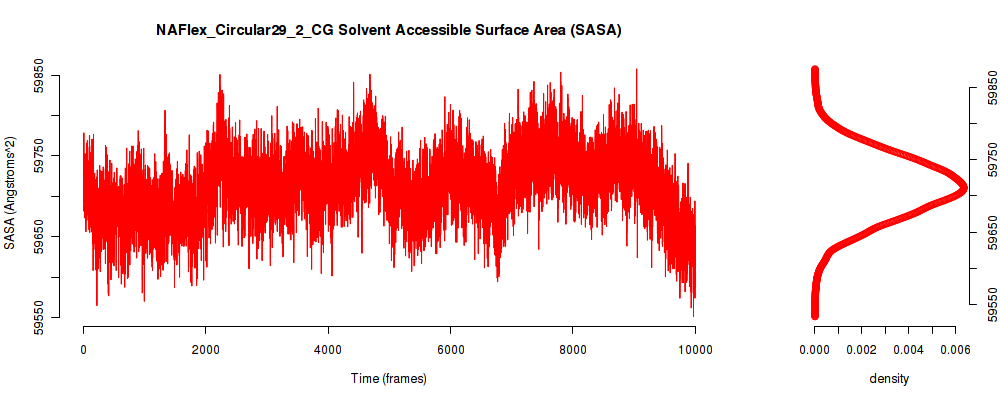
| SASA_avg | 59706.306 |
| SASA_stdev | 598.554 |
| SubType | B |
| Temperature | 298K |
| Topology | Circular29_2.pdb |
| Trajectory | Circular29_2.mdcrd |
| Water | No |
| _id | NAFlex_Circular29_2_CG |
| dataset | Array |
| date | 2024-Jul-08 |
| description | DNA-B Duplex Naked |
| forceField | CGeNArate |
| ionsModel | - |
| moleculeType | Dna |
| rev_sequence | - |
| saltConcentration | - |
| sequence | TCTCTCTCTCTCTCTCACCACCGCGTACAAGAAAGTTTGTTCTTTCATTTCGCCATACAGCTGCCTCGCGCGTTTCGGTGATATCGGTGAAAACCCTTCCCGTTTCGCTCAAGTTAGTATAAAAAAGCTGAACGAGAAACGTAAAATGATATAAATATCAATATATTAAATTAGATTTTGCATAAAAAACAGACTACATAATACTGTAAAACACAACATATGCAGTCACTATGAATCAACTACTTAGATGGTATTAGTGACCTGTAACAGAGCTGAGGCATCTTTCTCAATGAAAGTGTCGGAACTCGGTACATAGGCTTCCGGCTGATAGTAGTCGTATACGACTACTATCAGCCGGAAGCCTATGTACCGAGTTCCGACACTTTCATTGAGAAAGATGCCTCAGCTCTGTTACAGGTCACTAATACCATCTAAGTAGTTGATTCATAGTGACTGCATATGTTGTGTTTTACAGTATTATGTAGTCTGTTTTTTATGCAAAATCTAATTTAATATATTGATATTTATATCATTTTACGTTTCTCGTTCAGCTTTTTTATACTAACTTGAGCGAAACGGGAAGGGTTTTCACCGATATCACCGAAACGCGCGAGGCAGCTGTATGGCGAAATGAAAGAACAAACTTTCTTGTACGCGGTGGTGAGAGAGAGAGAGAGA |
| soluteAtoms | 678 |
| soluteCharge | - |
| soluteResidues | 678 |
| solventAtoms | - |
| solventResidues | - |
| time | 1,000 |
| totalAtoms | 678 |
| totalCharge | - |
| totalIons | - |
| totalResidues | 678 |