Entry: NAFlex_PYR
Nucleic Acid Data:
| Sequence | GGCGCC |
| Rev. Sequence | GGCGCC |
| Type | DNA |
| SubType | DNA unfolding |
| Chains | duplex |
| Pdb | - |
| Ligands | no |
| Keywords | DNA Duplex Naked ParmBSC1 TIP3P Electroneutral |
MD Simulation >> (Click to expand/shrink)
| Simulation Metadata | |
|---|---|
| Force Field | parmBSC1 |
| Simulation Date | 2015-Apr-29 |
| Simulated Time | 400 ns |
| Time Step | 1ps |
| Parts | ions + DNA |
| Temperature | 368K |
| Water | TIP3P |
| Additional Solvent | No |
| Counter Ions | Na+ |
| Ionic Concentration | Electroneutrality |
| Additional Ions | No |
| Additional Molecules | Pyridine |
| Ions Parameters | Dang |
Unified Molecular Modeling (UMM) Metadata
(Click to see full UMM)
(Click to see full UMM)
| Simulation Metadata (UMM) | |
|---|---|
| AdditionalIons | No |
| AdditionalMolecules | Pyridine |
| AdditionalSolvent | No |
| Author | A.P. |
| Chains | duplex |
| Comments | /orozco/scratch/parmBSC1/Alberto2/PYR |
| CounterIons | Na+ |
| Format | mdcrd |
| FrameStep | 1ps |
| Frames | 400000 |
| IonicConcentration | Electroneutrality |
| IonsParameters | Dang |
| Ligands | no |
| NucType | DNA |
| PDB | NONE |
| Parts | ions + DNA |
| RMSd_avg | 7.401 |
| RMSd_stdev | 3.142 |
| Rgyr_avg | 10.137 |
| Rgyr_stdev | 1.279 |
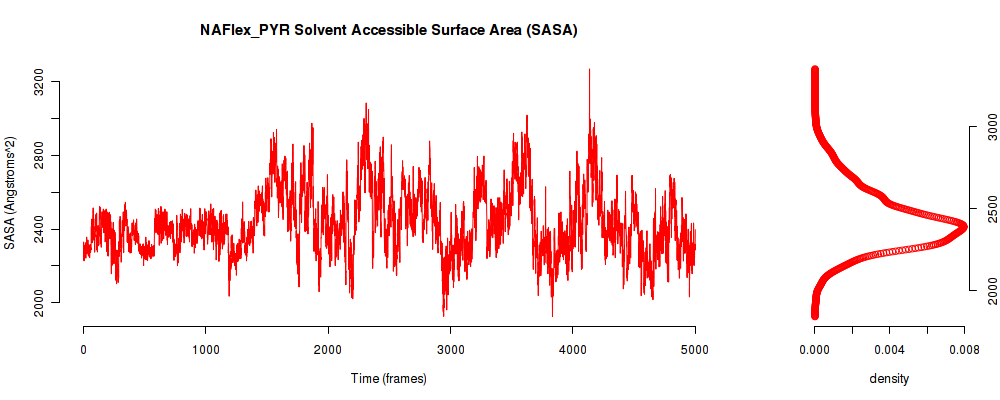
| SASA_avg | 2429.779 |
| SASA_stdev | 177.663 |
| SubType | DNA unfolding |
| Temperature | 368K |
| Topology | PYR.dry.top |
| Trajectory | PYR.trj.gz |
| Water | TIP3P |
| _id | NAFlex_PYR |
| authors | Ivan Ivani, Pablo D. Dans, Agnès Noy, Alberto Pérez, Ignacio Faustino, Adam Hospital, Jürgen Walther, Pau Andrio, Ramon Goñi, Alexandra Balaceanu, Guillem Portella, Federica Battistini, Josep Lluís Gelpí, Carlos González, Michele Vendruscolo, Charles A. Laughton, Sarah A. Harris, David A. Case and Modesto Orozco |
| boxAngle | 109.471219 |
| boxX | 45.6273186 |
| boxY | 45.6273186 |
| boxZ | 45.6273186 |
| dataset | Array |
| date | 2015-Apr-29 |
| description | DNA Duplex Naked ParmBSC1 TIP3P Electroneutral |
| forceField | parmBSC1 |
| ionsModel | - |
| moleculeType | Dna |
| rev_sequence | GGCGCC |
| saltConcentration | - |
| sequence | GGCGCC |
| soluteAtoms | 376 |
| soluteCharge | -10 |
| soluteResidues | 12 |
| solventAtoms | - |
| solventResidues | - |
| time | 400 |
| totalAtoms | 376 |
| totalCharge | -10 |
| totalIons | - |
| totalResidues | 12 |