Centre of Excellence for Biomolecular Research (BioExcel)

Life Science research has become increasingly digital, and this development is accelerating rapidly. Biomolecular modelling techniques have advanced tremendously.

Life Science research has become increasingly digital, and this development is accelerating rapidly. Biomolecular modelling techniques have advanced tremendously.
Multi-Scale Complex Genomics will provide tools to integrate the navigation in genomics data from sequence to 3D/4D chromatin dynamics data.
ParmBSC1 forcefield, a refined force-field for the atomistic simulation of DNA.

We are involved in a series of large scale biomedical and biotechnogical genomics projects, characterized by the need of manage large amounts of data. We provide data storage, mechanism for data transmission to project partners, and distribution of public data results. Advanced database both relational and noSql based ae available. Also we are adapting bioinformatics algorithms to large scale calculations compatible with the HPC environment.
Relevant projects

The project's objective is to develop new peptide labels (tags) and monoclonal antibodies (mAbs) specific for advanced tools for functional analysis of proteins in the areas of biotechnology and biomedicine. The structural design of tags from existing domains, the creation of structural variants by modelling, and preliminary analysis of its performance in vivo will allow planning and efficient collection and analysis of monoclonal antibodies.

We develop coarse-grained models with different levels of resolution and enhanced sampling algorithms to study efficiently large scale protein conformational transitions, protein aggregation, macromolecular assemblies and protein-protein interactions in very large systems.
Large systems and long timescales that would demand an unfeasible simulation time with usual atomistic molecular dynamics methods become feasible when using coarse-grained representations of the system with a lower resolution than the atomistic representation used in standard molecular dynamics simulations.

Protein dynamics is the key to understand biological function. We are interested in the development of coarse-grained simulation techniques based on Elastic Network Models that allow to deal with large molecular systems and extended time scales.
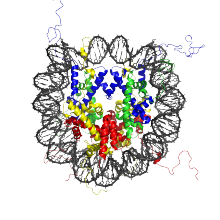
We focus on genome-wide regulatory mechanisms signaled by the underlying DNA structure, identifying distinctive physical properties that might affect nucleosome positioning in key regulatory regions like transcription start and termination sites, and understanding how these factors modulate gene expression.

Epidermal Growth Factor Receptor is a membrane Tyrosine Kinase and acts as a major oncogene in a number of tumors. We are studying the conformational changes of the extracellular domain using a combination of submicrosecond Molecular Dynamics simulations and Elastic Network Models.

MoDEL (Molecular Dynamics Extended Library) is a database holding over 1,800 trajectories of 10ns length from a representative set of protein structures. From the original MoDEL database, a set of interesting simulation libraries have been derived and extended: nMoDEL, µMoDEL, kMoDEL and pMoDEL.