NAFlex Coarse-Grained DNA algorithms
NAFlex provides a couple of Coarse-grained algorithms to obtain a dynamic view of a DNA molecule in a lower resolution:
DNA Elastic Mesoscopic Model (Base level)
The Elastic Mesoscopic Model represents an intermediate level of resolution (base pairs) and potential energy complexity. It assumes that DNA deformations can be approximated as the addition of harmonic distortions of equilibrium base-pair step geometries. Three rotational (twist, roll and tilt) and three translational (slide, shift and rise) degrees of freedom are considered, which allows us to define the Hamiltonian as:
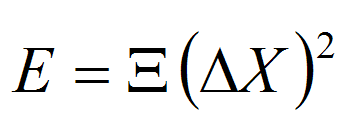
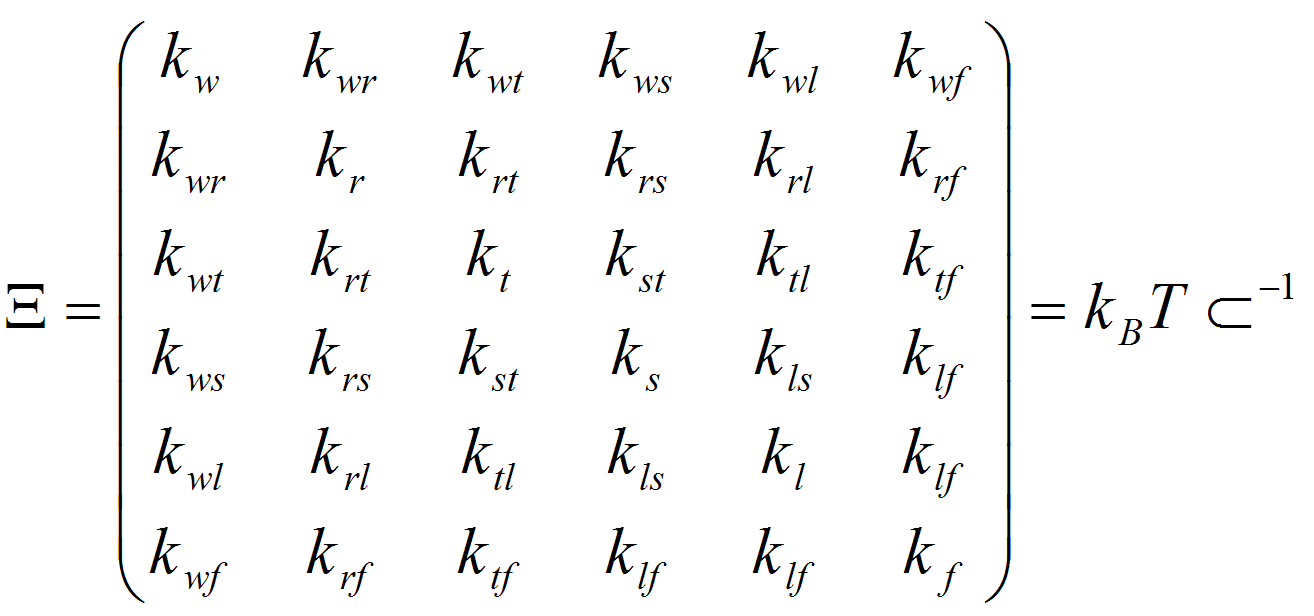
where kB is the Boltzman constant, T is the absolute temperature, E is the energy associated to the deformation ΔX, and k stands for the different stiffness constants defined by the 36 elements of the stiffness matrix (Ξ): Twist (w), Roll (r), Tilt (t), Rise (s), Slide (l) and Shift (f). The stiffness matrix (Ξ) can be obtained by the inversion of the covariance matrix generated from either analysis of Molecular Dynamics trajectories or from the analysis of dinucleotide step variability in the crystal structures of DNAs and DNA-Protein complexes. In all cases samplig was obtained using MonteCarlo simulations in the helical coordinate space following DNAlive protocol.
Once a movement in helical coordinates space is accepted by Metropolis test, the corresponding Cartesian structure of the fiber is generated, thus finally obtaining a trajectory for a later analysis with NAFlex and/or video visualization using JMol java scripts.
DNA Worm-like Chain Model (one bead x M base pair steps)

NAFlex implements a Worm-Like Chain (WLC) Montecarlo algorithm to obtain a coarse-grained DNA dynamics.
In Worm-Like Chain Model, the DNA is interpreted at the lowest level of resolution. The combined Worm-Like chain (WLC) model ( Jian et al, J.Comp.Phys. 136, 168-179 ) includes a mechanical and an electrostatic potential. In this treatment, a DNA segment is represented as an elastic chain of N beads, each bead comprising M base pairs (one bead x M base pair steps), connected by N-1 inter-bead segments of average length l0=1.36 nm. To account for the torsional deformation, a local coordinate system is fixed to each DNA bead, and a set of Euler angles { αi, βi, γi } that transform from the coordinate frame of bead i to that of bead i+1 are defined.
The total potential energy is a sum of stretching, bending, torsion, and electrostatic contributions:
E = ES + EB + ET + EDH
The stretching energy ES, is specified by:
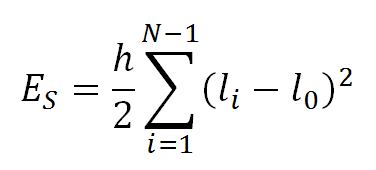
where li is the length of segment i and h is the stretching force constant. The value of h was set to 100 KBT/l02 by Jian et al., where KB is the Boltzmann constant and T the temperature, as it reproduces the correct DNA bond variance.
The bending energy, EB, is given by:
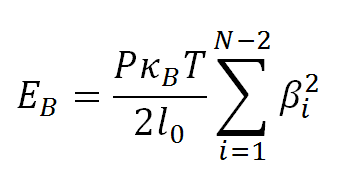
where βi is the Euler bending angle of segment i relative to segment i+1, and P=50 nm-1 is the DNA bending persistence length.
The torsional energy, ET, is computed as:

where C=2×10-19 erg∙cm is the consensus value for the DNA torsional rigidity constant, the sum of αi and γi Euler angles defines the torsion between the local coordinate systems of two consecutive beads, and the equilibrium angle ϕ0 is a parameter that gives the mean DNA twist and depends on the bead-bead equilibrium distance and the helical repeat.
Finally, the electrostatic energy, EDH, is modeled using the Debye-Hückel potential that accounts for screening by monovalent ions in solution:
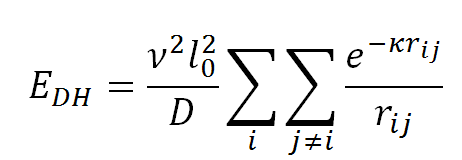
where v is the salt-dependent effective DNA linear charge density, D is the dielectric constant of water, rij is the distance between the centers of beads i and j, and k is the inverse Debye length. As done in Jian et al., the parameter v is calculated through the procedure developed by Stigter (Stigter, D., 1977, Biopolymers, 16, 1435-1448) to treat DNA electrostatics, in which the solution to the Poisson-Boltzmann equation for an infinitely long and uniformly charged DNA cylinder is approximated to a solution of the Debye-Hückel equation for a uniform charge density.
Equilibrium phase-space sampling is achieved through a Metropolis Monte Carlo procedure. At each step, a pivot DNA bead and an axis passing through it are selected at random, and one of the following trial moves is attempted:
- (a) local rotation or translation with respect to the chosen pivot axis;
- (b) global rotation of the shorter part of the chain (i.e., from the pivot DNA bead to either the first or last bead) through the chosen pivot axis.
The two moves are uniformly distributed in ranges { –δ, +δ } and the values of δ are determined so that the acceptance ratio lies around ~30-35%.