Welcome to Nucleosome Dynamics
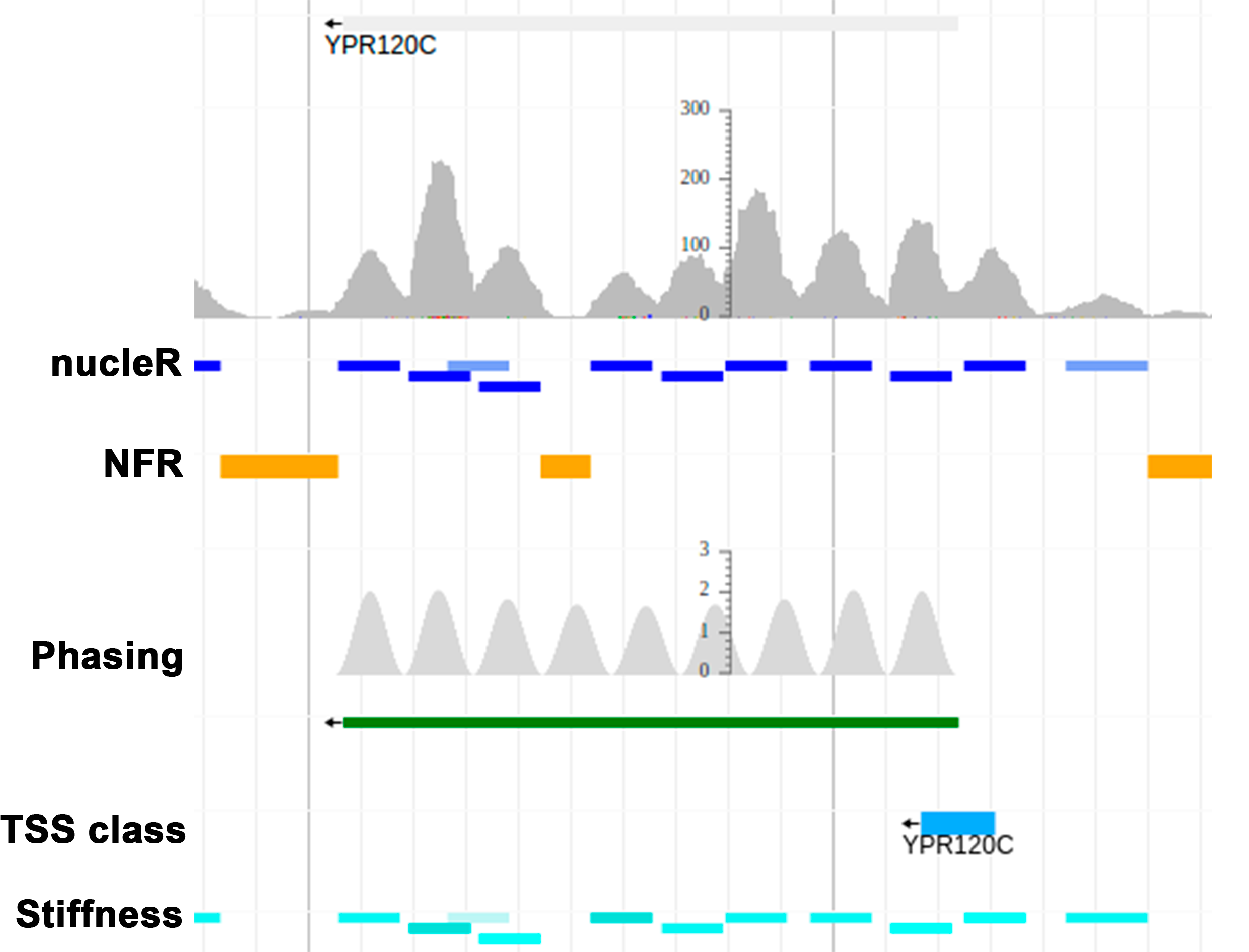
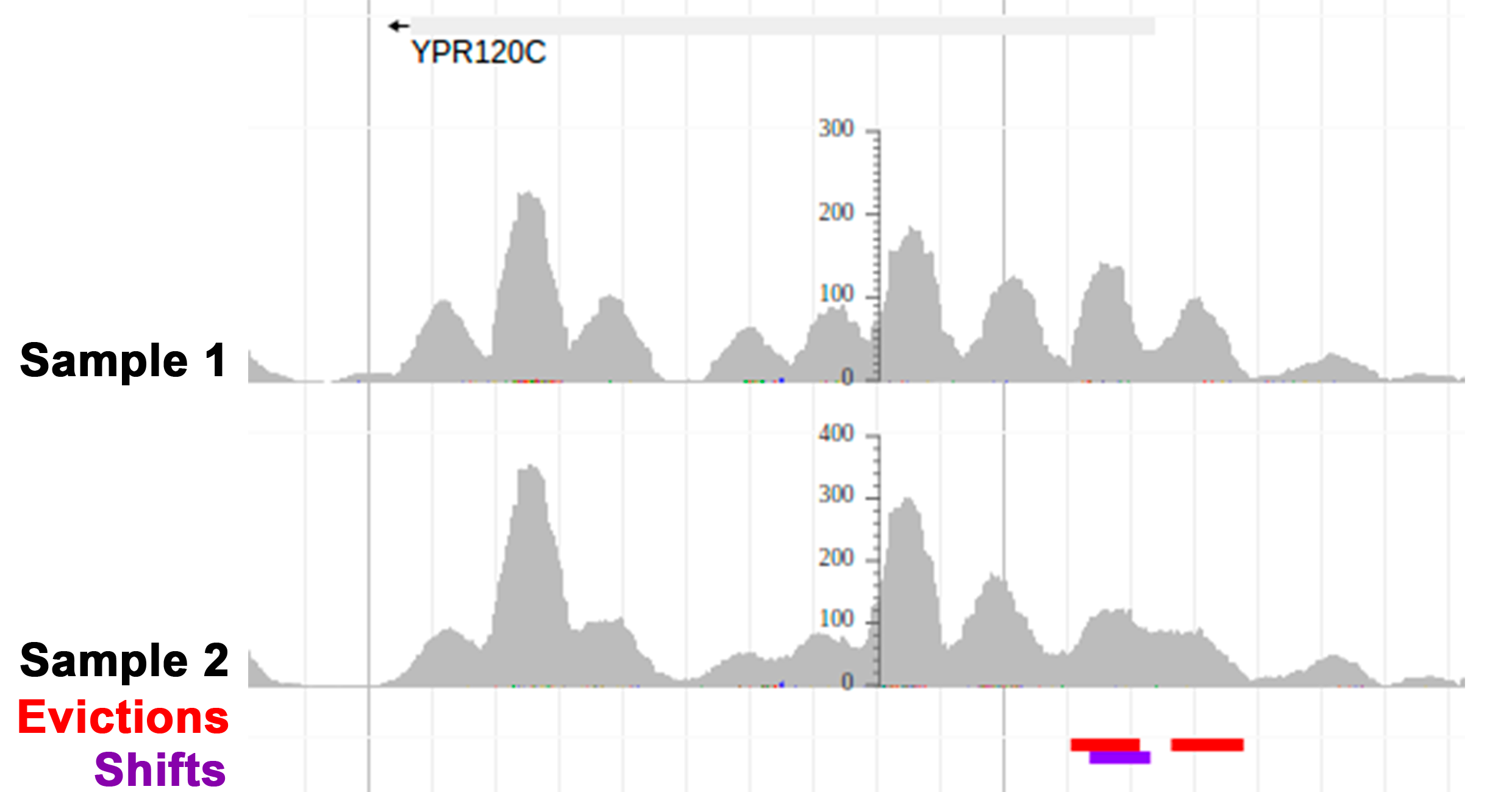
Nucleosome positioning plays a major role in transcriptional regulation and most DNA-related processes. The Nucleosome Dynamics server offers different tools to analyze nucleosome positioning from MNase-seq experimental data and to compare experiments to account for the transient and dynamic nature of nucleosomes under different cellular states.
Two specific programs, nucleR and NucDyn, were specifically developed to perform such studies.
- nucleR performs Fourier transform filtering and peak calling, in order to efficiently and accurately define and classify the location of nucleosomes.
- NucDyn is a method to detect changes in nucleosome architectures based on MNase-seq experiments. It identifies nucleosomes’ insertions, evictions and shifts between two experiments at the read level.
Additionally, a list of other nucleosome related features:
- Location of nucleosome-free regions
- Classification of transcription start sites based on the surrounding nucleosomes
- Study of nucleosome periodicity at gene level
- Stiffness of the nucleosomes derived from fitting a Gaussian function to nucleosome profiles
From this page you can access the MuGVRE and Galaxy framework, where you can upload, analyse and visualize your MNase-seq data. You can also find the link to nucleR and NucDyn as open source R/Bioconductor packages to install and execute on your computer.
- 2019-12-10 Our recent publication in NAR (https://dx.doi.org/10.1093/nar/gkz759) has been highligted by the Biophysical Society of Spain as a paper of the month in the Biofísica Magazine https://biofisica.info/topics/highlighted-papers/highlights2019/oct019/
- 2019-08-31 Publication already online
- 2018-09-17 3DAROC18: 3C-data Analysis and 3D Chromatin Folding
- 2017-12-13 Multi-scale study of 3D Chromatin structure – 2nd edition
- 2017-11-15 Multi-scale study of 3D Chromatin structure
- 2017-04-10 Multi-scale study of 3D Chromatin structure
- 2016-10-10 3DAROC16: 3C-based data analysis, 3D reconstruction of chromatin folding and Nucleosome Dynamics
Diana Buitrago , Laia Codó , Ricard Illa, Pau de Jorge, Oscar Flores, Federica Battistini, Genís Bayarri, Romina Royo, Marc Del Pino, Simon Heath, Adam Hospital, Josep Lluís Gelpí, Isabelle Brun Heath and Modesto Orozco. 2019. “Nucleosome Dynamics: a new tool for the dynamic analysis of nucleosome positioning.” Nucleic Acids Research, gkz759. https://doi.org/10.1093/nar/gkz759
Molecular Modeling and Bioinformatics Group
BSC Computational Bioinformatics Node








