Nucleosome Dynamics Help - Usage
Usage
How to run Nucleosome Dynamics
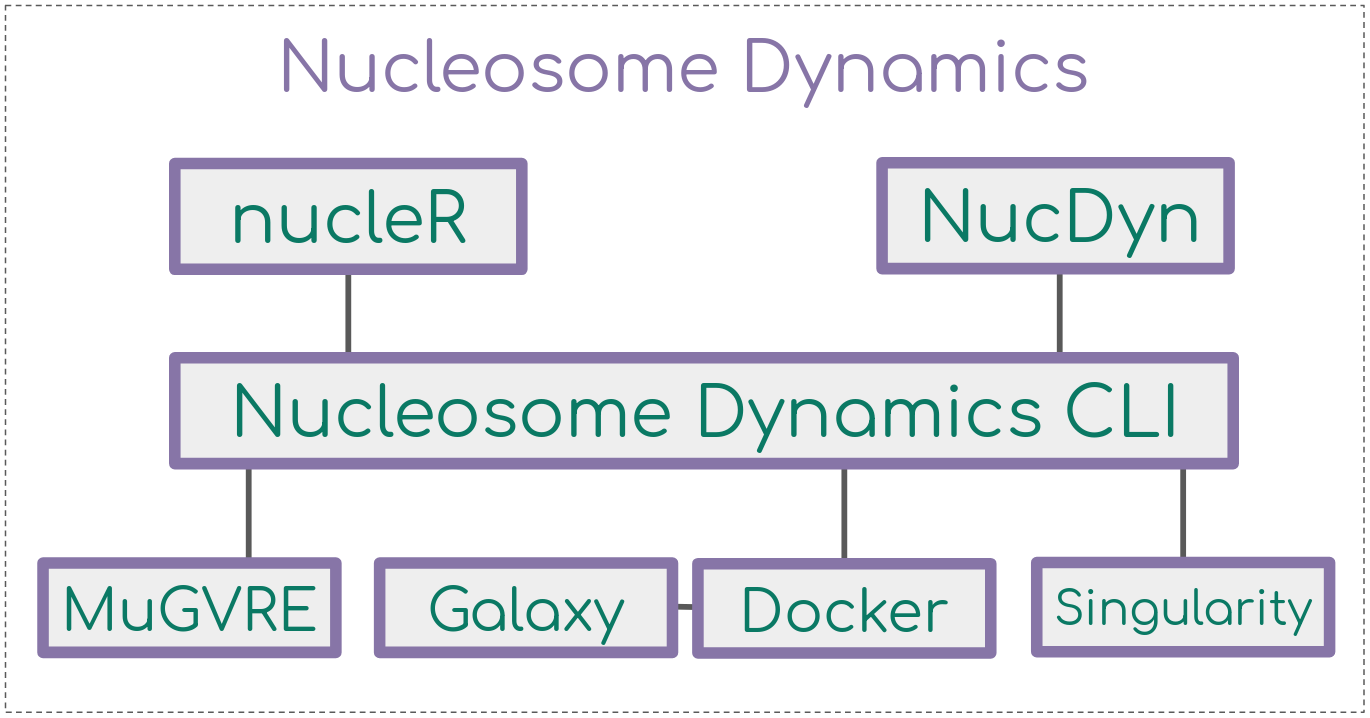
Nucleosome positioning plays a major role in transcriptional regulation DNA replication and DNA repair. The suite is written in R, yet, a number of other implementations have been prepared in order to reach also non-R users. These implementations include two web-based platforms (MuGVRE and Galaxy) and two containerized installations (docker and singularity).
Learn how to run Nucleosome Dynamics analyses in each of the offered distributions: