----
MDWeb interface
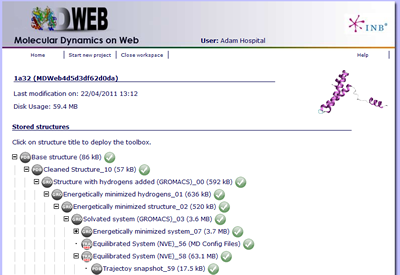 MDWeb holds a personal workspace where structures and trajectories are stored.
MDWeb holds a personal workspace where structures and trajectories are stored.
Data is structured in "projects". A project is opened with an initial structure or trajectory. All operations are based on that initial input.
Data in the project is organized in a tree that outlines the history of every object back to the initial one.
Structures and trajectories are added to the tree once available.
The available operations and workflows are defined according to every data type.
User Registration
 MDWeb gives the possibility to work as an anonymous user, or as a registered user.
MDWeb gives the possibility to work as an anonymous user, or as a registered user.
Warning: Anonymous users' projects will be removed once disconnected or when session expires (after some minutes of inactivity).
On the other hand, Registered users' projects will be stored in our disks for a reasonable time. A capacity of 2 GB of disk space will be assigned for each registered user.
New Project
MDWeb offers three main entry options:
- Simulation (Single Structure): Work from a structure, setup and run an MD simulation, run a coarse-grained MD.
- Analysis (MD Trajectory): Work from a trajectory, analyse, get information or convert between MD trajectory formats.
- DNA/RNA Simulation from Sequence: Work from a sequence, obtain a 3D structure from a nucleic acid sequence.
- Upload past MDWeb Project: Upload a stored MDWeb project.
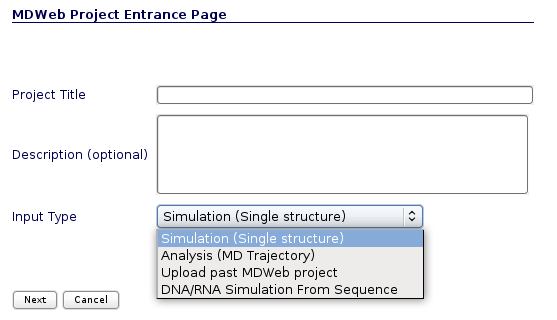
If Simulation (Single Structure) option is chosen, user will be redirected to the Structure Checking page.
(*) Please note that user provided structures must follow PDB File Format v.3.30 (July 13, 2011). The most relevant points to consider are:
- A Non-blank alphanumerical character is used for chain identifier.
- Non-polymer or other “non-standard” chemical coordinates, such as water molecules or atoms presented in HET groups, use the HETATM record type.
Note that MDWeb has a limit file size of 100 MB.
Different input files are required depending on program and trajectory formats (NAMD/Amber/Gromacs). Gzipped files are accepted.
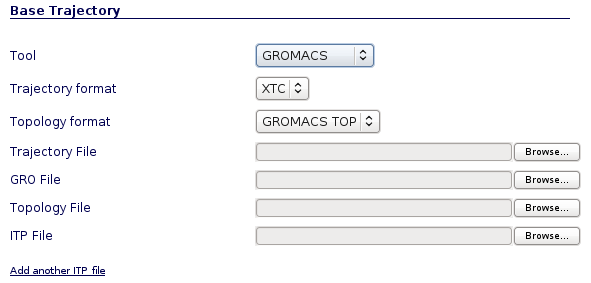
Gromacs Trajectory Input Files Example
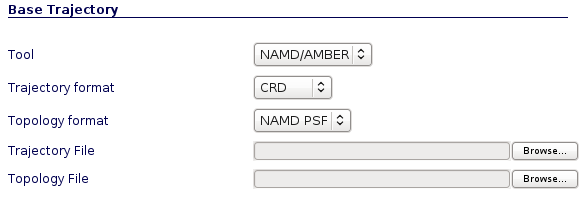
NAMD/Amber Trajectory Input Files Example
The Upload past MDWeb Project option allows MDWeb users to upload projects, previously strored using the Download Project utility.
Upload Project
The DNA/RNA Simulation from Sequence option allows MDWeb users to upload a nucleic acid sequence, directly or from FASTA file, in order to obtain a 3D structure. User have different kind of final structures to choose:
- Right Handed B-DNA
- Left Handed B-DNA
- Right Handed A-DNA
- Right Handed A-RNA
- Right Handed A'-RNA
- User Defined DNA/RNA: Possibility to change a set of helical parameters.
- DNA generated using ABC average helical parameter values. Sequence-dependent: each base pair step is assembled using corresponding average helical parameters.
- DNA generated using experimental X-ray average helical parameter values. Sequence-dependent: each base pair step is assembled using corresponding average helical parameters.
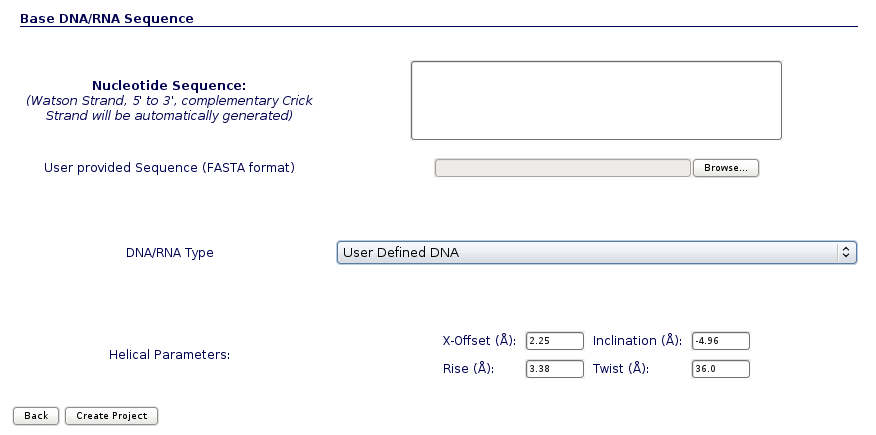
Input from Sequence