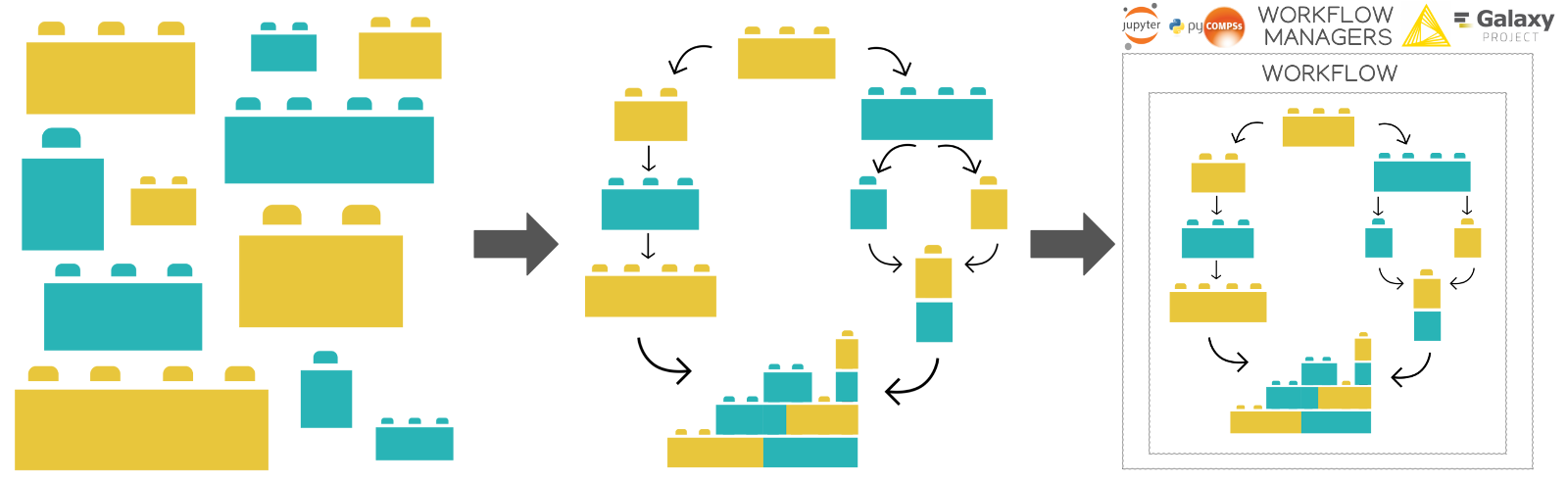
BioExcel Building Blocks, a software library for interoperable biomolecular simulation workflows
 BioExcel Building Blocks
BioExcel Building Blocks

Help us to improve!
Please help us to improve the BioExcel Building Blocks by filling in our 2023 survey.
- 22/11/2021
24/11/2021 BioExcel Virtual Training #3 - Virtual Trainings - 09/11/2020
13/11/2020 BioExcel Virtual Training #2 - Virtual Trainings - 23/10/2020 BioExcel Virtual Training AZ - Virtual Trainings
- 16/09/2020 BioExcel Webinar #48 - Webinars
- 23/05/2023 BioExcel Webinar #69 - Webinars
- 29/05/2024 BioExcel Webinar #78 - Webinars
- 04/09/2020 Pre Exascale Tutorial - Tutorials
- 23/06/2020 Summer School 2020.1 - Workshops
- 23/06/2020 Summer School 2020.2 - Workshops
- 08/06/2021 Summer School 2021.1 - Workshops
- 08/06/2021 Summer School 2021.2 - Workshops
- 13/06/2022 Summer School 2022 - Workshops
- 11/09/2023 Summer School 2023 - Workshops
- 08/02/2022 CSC 2022 - Workshops
- 09/12/2019
12/12/2019 BioExcel Virtual Training #1 - Virtual Trainings - 02/02/2021 CSC / BioExcel workshop - Workshops
- 27/05/2024
2024.1
version
- New biobb_haddock package, a module collection to compute information-driven flexible protein-protein docking.
- New biobb_pytorch package, a module collection to create and train ML & DL models using the popular PyTorch Python library.
- New Docker images and Dockerfiles for all BioBB workflows.
- Using interactive Jupyter Notebooks and BioConda for FAIR and reproducible biomolecular simulation workflows paper released.
- Now all BioBB workflows are available in Google Colab.
- All BioBB's updated to 4.2.0+ version.
- 04/12/2023
2023.4
version
- New biobb_pdb_tools package, a swiss army knife for manipulating and editing PDB files.
- All BioBB workflows updated to latest version.
- 08/09/2023
2023.3
version
- All BioBB's updated to 4.1.0+ version.
- Update to Python 3.8 or greater.
- Biopython is no longer linked to a fixed version.
- Major fixes in biobb_common.
- 27/06/2023
2023.2
version
- New demonstration workflow: FAIR workflows to chart and characterize the conformational landscape of native proteins (10.7490/f1000research.1119506.1).
- BioBB-Wfs and BioBB-API webinar.
- biobb_pmx updated to the last PMX version, that accepts alchemical ligand modifications.
- 13/04/2023
2023.1
version
- All BioBB's updated to 4.0.0+ version.
- Major fixes in biobb_common.
- New biobb_flexdyn package, a module collection for studies on the conformational landscape of native proteins.
- New biobb_flexserv package, a category for biomolecular flexibility studies on protein 3D structures.
- New biobb_godmd package, a module collection to compute protein conformational transitions with the GOdMD method.
- 28/12/2022
2022.4
version
- All BioBB's updated to 3.9.0+ version.
- Major fixes in biobb_common.
- biobb_md has been discontinued, superseeded by biobb_gromacs.
- 25/06/2022
2022.2
version
- New biobb_cp2k package, a module collection to allow setup and simulation of QM simulations using CP2K QM package.
- New tools for biobb_io and biobb_structure_utils.
- 17/03/2022
2022.1
version
- Extended information in Python Docstrings improving workflow language interoperability.
- Released new BioBB Workflows website.
- 24/12/2021
2021.4
version
- Major fixes in biobb_common.
- Released new BioBB REST API.
- 20/10/2021
2021.3
version
- All BioBB's updated to 3.7.0+ version.
- New biobb_dna, a package composed of different analyses for nucleic acid trajectories.
- New ABC MD Setup workflow.
- New Structural DNA helical parameters workflow.
- 30/06/2021
2021.2
version
- All BioBB's updated to 3.6.0+ version.
- Update to Biopython 1.79.
- New biobb_cmip package, a module collection to compute classical molecular interaction potentials.
- New set of Protein-Ligand Docking workflows.
- New set of AMBER workflows.
- 10/03/2021
2021.1
version
- New biobb_vs package, a module collection to perform virtual screening studies.
- New biobb_amber package, a category for AMBER MD package.
- 25/12/2020
2020.4
version
- All BioBB's updated to 3.5.0+ version.
- Update to Biopython 1.78.
- New extended and improved JSON schemas (Galaxy and CWL-compliant).
- New MemProtMD DB REST API tools in biobb_io.
- New Resampling module in biobb_ml.
- New biobb_md grompp_mdrun block combination.
- New "shell" option in biobb_md Solvate.
- New biobb_model tools.
- Biobb_model now is using Modeller.
- 15/10/2020
2020.3
version
- Dependency biobb_common has been updated to 3.0.1 version.
- PMX software has been updated to python 3 and thus, biobb_pmx breaks its dependence from docker.
- 16/06/2020
2020.2
version
- All BioBB's updated to 3.0.0+ version.
- Update to Python 3.7.
- Update to Biopython 1.76.
- New biobb_ml package, a module collection to perform machine learning predictions.
- New workflow to perform free energy calculations over mutated structures (PMX).
- 05/03/2020
2020.1
version
- All BioBB's updated to 2.0.0+ version.
- Compatibility with software containers (Docker, Singularity).
- New documentation, tutorials and website.
- 20/11/2019
2019.4
version
- All BioBB's updated to 1.0.2+ version.
- New adapters for PyCompSs, CWL and Galaxy.
- New workflows for MD and protein mutations.
BioExcel
Building Blocks, a software library for interoperable
biomolecular simulation workflows.
Pau Andrio,
Adam Hospital, Javier Conejero, Luis Jordá, Marc Del Pino,
Laia Codo, Stian Soiland-Reyes, Carole Goble, Daniele Lezzi,
Rosa M. Badia, Modesto Orozco & Josep Ll. Gelpi Nature
Scientific Data, 09/2019, Volume 6, Issue 1, p.169,
(2019)

BioExcel is funded by the European Union Horizon 2020 program under grant agreements 823830, 675728.