BioExcel Building Blocks - Success Stories
Success Stories
The BioExcel Building Blocks library is currently being used in the BioExcel project to build and run biomolecular simulation workflows for a set of defined scientific use cases. A couple of studies from these use cases are presented as examples of success stories showing what can be done using the library.
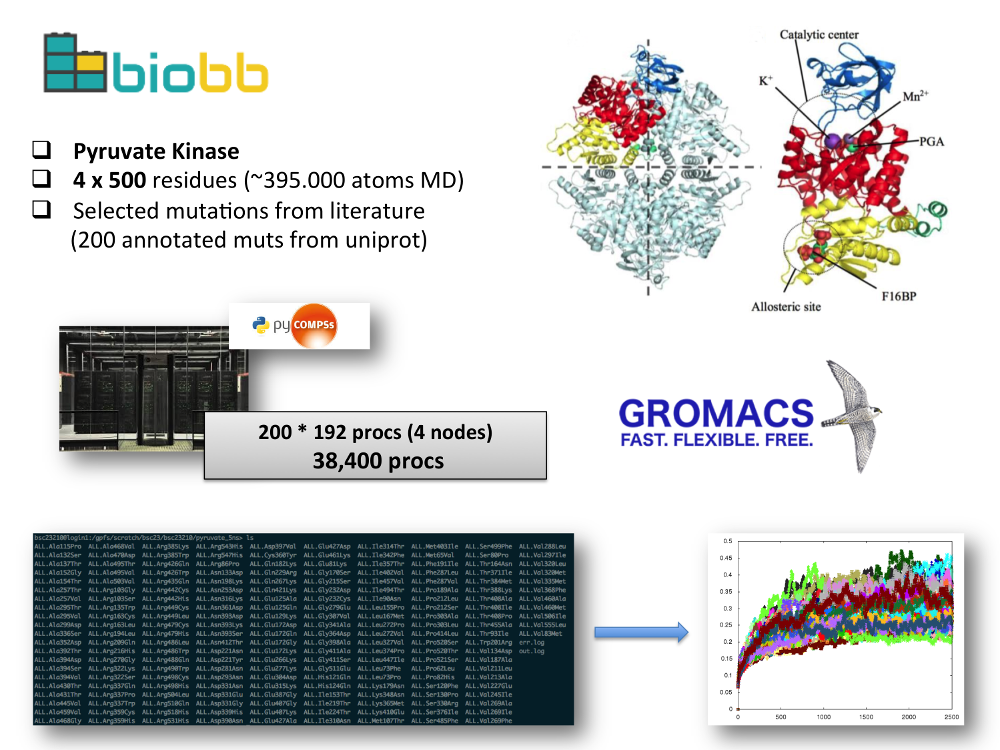
- Mutations effect on the protein structural flexibility: study of differences in structural flexibility caused by a set of annotated variants on different protein families. A test set was run using the Pyruvate Kinase protein. Observations such as structural and dynamics features extracted from MD trajectories are used in the context of prediction of the pathogenicity of such variants, coupling structure and flexibility with function.
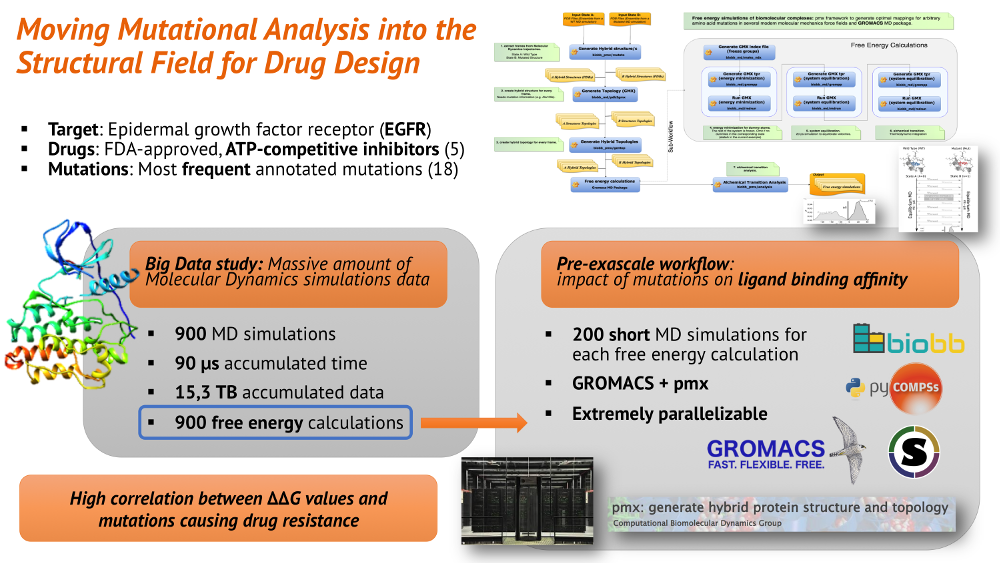
- Identification of mutations causing drug resistance from ligand binding free energies: study of the effect caused by a set of annotated mutations on the ligand binding by means of MD simulations and free energy calculations. A test set was run using the Epidermal Growth Factor Receptor (EGFR) protein. Observations suggest a high correlation between ΔΔG values and mutations causing drug resistance.
Mutations effect on the protein structural flexibility
- Test system: human erythrocyte pyruvate kinase, was selected because of its large number of annotated missense variants that have been associated to a disease called nonspherocytic hemolytic anemia, a rare, autosomal recessive disease which causes blood disorders characterized by the premature destruction of red blood cells (erythrocytes), resulting in anemia. A set of 200 mutations consisting on reported pathogenic variants were manually selected and curated from the whole set of variants available at the UniprotKB database.
- Workflow: Python script looping over the 200 mutations, modeling the mutation on the initial structure (Pyruvate Kinase WT), running a complete MD setup process (adding and minimizing hydrogen atoms energy, adding solvent box and counterions, equilibrating the energy of the system) and finally running the MD simulation using GROMACS MD package. The workflow is controlled by PyCOMPSs workflow manager, and an execution over 38,400 cores was successfully run in the Marenostrum supercomputer (BSC), obtaining information from 200 independent MD simulations in just 24h. Modules used: biobb_io, biobb_model, biobb_md, biobb_analysis.
Identification of mutations causing drug resistance from ligand binding free energies
- Test system: EGFRs are transmembrane receptors located on the cell membrane. EGFRs play an important role in controlling normal cell growth, apoptosis and differentiation. Mutations of EGFRs can lead to abnormal activation and signal transduction causing unregulated cell division and ultimately driving some types of cancers. A therapeutic approach is knocking down the intracellular tyrosine-kinase activity. A set of approved small molecule drugs exists, with identified mutations leading to resistance. The study of the correlation of these mutations with the resistance observed will open the door to new therapeutic strategies.
- Workflow: Python script performing a free energy calculation using GROMACS and pmx packages. The fast growth thermodynamic integration approach is used (see pmx tutorial). Taking a couple of independent equilibrium MD simulations as input (apo + holo), the pipeline builds and runs 100 forward and 100 reverse short MD simulations, and finally collects all results to compute the ligand binding free energy estimation. The workflow is controlled by PyCOMPSs workflow manager, which exploits the inherent concurrency of the pipeline. Modules used: biobb_md, biobb_pmx, biobb_analysis.