BioExcel Building Blocks software library
The BioExcel Building Blocks (biobb) software library is a collection of Python wrappers on top of popular biomolecular simulation tools.
The library offers a layer of interoperability between the wrapped tools, which make them compatible and prepared to be directly interconnected to build complex biomolecular workflows.
All the building blocks share a unique syntax, requiring input files, output files and input parameters (properties), irrespective of the program wrapped.
The available building blocks are divided in categories (biobb modules) depending on their functionalities and tools wrapped.
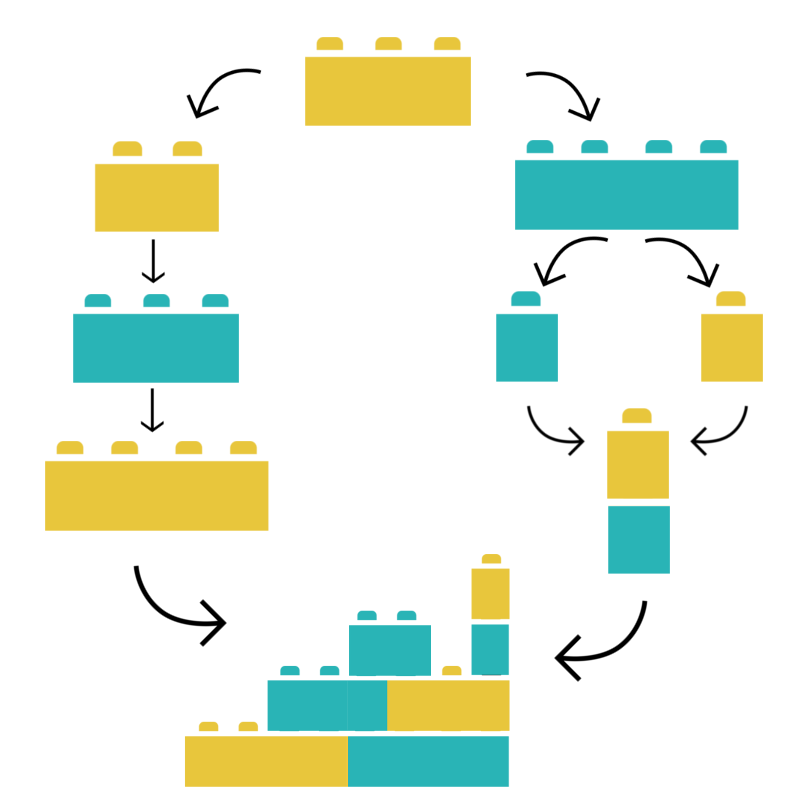
Building and sharing complex biomolecular simulation workflows just require joining and connecting BioExcel Building Blocks together. Workflows built up from the biobb library are:
- Reusable & Reproducible: A workflow can be packaged in a single Python script with a defined Conda environment, or in a CWL specification file, to be shared and reproduced anywhere else.
- Easy to build, using Drag & Drop GUIs (Galaxy, KNIME), easy to develop and test, using interactive GUIs (Jupyter Notebooks), and easy to share, using packaging tools and standards for describing analysis workflows (Conda environments, CWL specifications).
- Prepared for the exascale, thanks to the adapter interoperability layer and the compatibility with HPC-ready workflow managers such as PyCOMPSs or Toil.
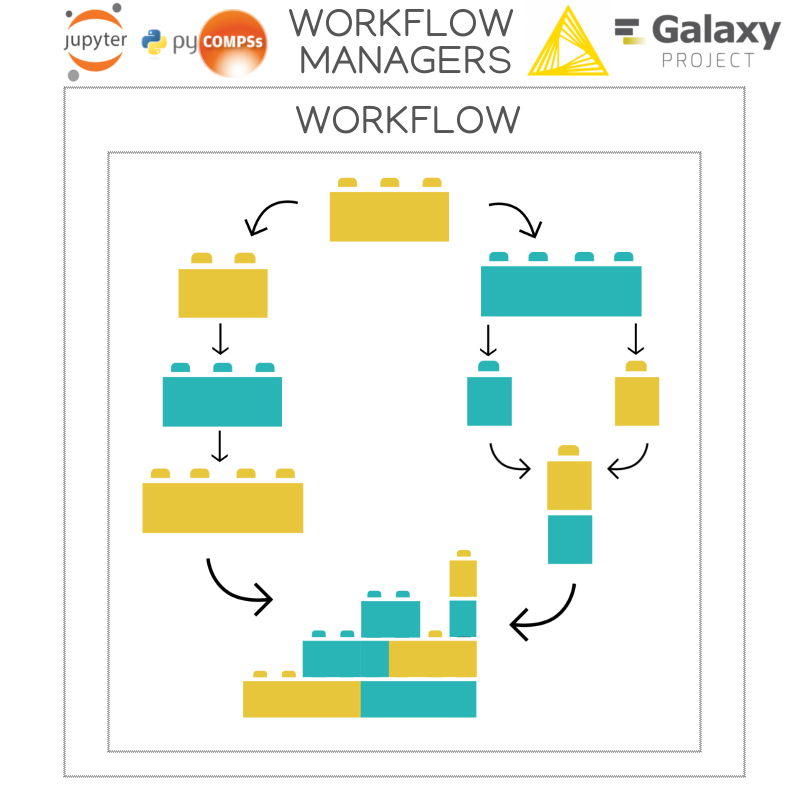
Choose a workflow manager and an infrastructure to launch your biomolecular simulation workflows.
Packaging of the library modules allow easy installation and run in many different infrastructures: Desktop, VM, Cloud, HPC supercomputers, etc.
An adapter interoperability layer allow the control of the workflow executions using different workflow managers.
Compatible workflow managers cover very distinct areas:
- GUI, Interactive: Jupyter Notebooks, Web servers
- GUI, Drag & drop: Galaxy, KNIME*
- HPC: PyCOMPSs, Toil, CWLexec
* Work in progress
