BIGNASim database structure and analysis portal for nucleic acids simulation data
----
DNA & RNA Structure and Helical Parameters Analyses (NAFlex)
----Tutorial 2 -- Global analysis (XCGY)
Tutorial 3 -- Meta-trajectory (XCGY)
Tutorial 4 -- Experimental vs MD analysis
----Experimental Average Values
In order to compare global analysis results stored in our database coming from MD simulations with experimental values, we have built a complex pipeline to obtain such values.
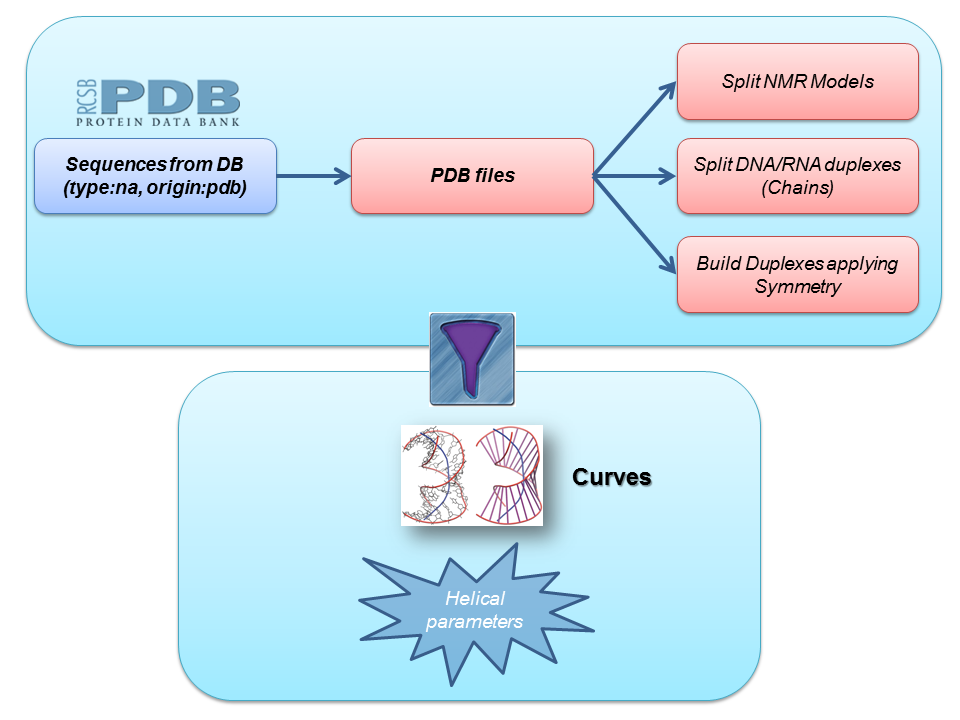
Starting from all the PDB files classified as Nucleic acid, we built a complete dataset of duplex structures following these steps:
- Splitting NMR models in different files
- Identifying DNA/RNA chains forming duplexes and extracting them to different files
- Applying symmetry to those PDB files that need a transformation to obtain the final biomolecule (see PDB REMARK 350)
Next step in the pipeline consists in applying a list of filters to the set of duplex nucleic acid structures obtained. These filters work as a pre-process to the analyses we are interested in, and include, for example, checking for different strand lenghts, possible sequence mismatches, presence of essential atoms needed for subsequent analyses, etc. The final step consists in running Curves+ program to extract helical parameters information from the generated PDB files.
As an example of what we obtained, the following table shows the average results for the base-pair steps helical parameters computed for the complete list of experimental structures, together with values extracted from a similar previous work in the group (P. Dans et. al, NAR 2012 40, 10668-10678).
- Amount of data(N), average values and standard deviation of the six intra-strand base pair parameters for the 10 unique bps.
- For each step, the naked-DNA structures (first row), PDB structures (second row), and results from MD simulations with parmbsc0 (third row) from the paper cited are shown together with our new values computed to the set of filtered PDB files (forth row).
- Values for simulations were obtained from time averages computed for individual steps in each sequence.
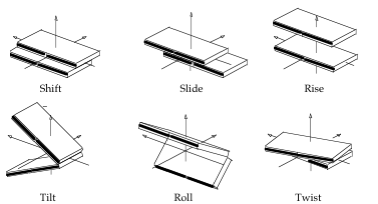
- Rotational parameters are in degrees (Twist, Tilt, Roll), and translational ones (Shift, Slide, Rise) in Å.
- Complementary steps (e.g. AG and CT) have the same average except for a change in sign of Shift and Tilt, and the same standard deviation.
| Step | Amount of data | Twist | Tilt | Roll | Shift | Slide | Rise |
| AA | 60 | 35.6±4.3 | -1.4±2.7 | 1.0±3.8 | 0.0±0.3 | -0.2±0.4 | 3.3±0.1 |
| AA | 850 | 33.4±5.9 | -1.2±3.0 | 2.5±7.1 | 0.1±0.5 | 0.0±0.6 | 3.3±0.2 |
| AA | 1M | 35.4±5.4 | -2.6±4.1 | 0.3±5.6 | -0.3±0.6 | -0.3±0.6 | 3.3±0.3 |
| AA | 4331 | 34.2±8.1 | -1.2±6.0 | 1.6±10.0- | 0.1±0.6 | -0.1±0.7 | 3.3±0.5 |
| TT | 60 | 35.6±4.3 | -1.4±2.7 | 1.0±3.8 | 0.0±0.3 | -0.2±0.4 | 3.3±0.1 |
| TT | 850 | 33.4±5.9 | -1.2±3.0 | 2.5±7.1 | 0.1±0.5 | 0.0±0.6 | 3.3±0.2 |
| TT | 1M | 35.4±5.4 | -2.6±4.1 | 0.3±5.6 | -0.3±0.6 | -0.3±0.6 | 3.3±0.3 |
| TT | 3154 | 34.3±7.5 | 0.9±7.6 | 1.8±10.1 | 0.1±0.6 | -0.2±0.7 | 3.3±0.5 |
| AC | 12 | 35.6±2.3 | -0.5±2.2 | 0.4±3.6 | -0.1±0.6 | -0.2±0.3 | 3.3±0.1 |
| AC | 670 | 31.5±3.9 | 0.7±3.4 | 2.0±4.1 | 0.2±0.6 | -0.6±0.4 | 3.2±0.2 |
| AC | 1M | 32.0±5.1 | -0.7±4.0 | -0.6±5.9 | 0.1±0.7 | -0.6±0.5 | 3.3±0.3 |
| AC | 2767 | 31.2±12.8 | -0.7±11.6 | 0.7±10.4 | 0.0±1.0 | -0.4±1.1 | 3.4±0.7 |
| GT | 12 | 35.6±2.3 | -0.5±2.2 | 0.4±3.6 | -0.1±0.6 | -0.2±0.3 | 3.3±0.1 |
| GT | 670 | 31.5±3.9 | 0.7±3.4 | 2.0±4.1 | 0.2±0.6 | -0.6±0.4 | 3.2±0.2 |
| GT | 1M | 32.0±5.1 | -0.7±4.0 | -0.6±5.9 | 0.1±0.7 | -0.6±0.5 | 3.3±0.3 |
| GT | 2683 | 30.8±9.2 | 0.1±10.9 | 0.6±9.7 | -0.1±0.7 | -0.4±0.8 | 3.3±0.7 |
| AG | 17 | 29.7±5.9 | -0.4±4.0 | 5.7±4.9 | 0.4±0.4 | 0.3±0.8 | 3.4±0.3 |
| AG | 700 | 32.4±4.7 | -1.1±3.6 | 3.2±5.7 | 0.1±0.7 | -0.2±0.3 | 3.3±0.3 |
| AG | 1M | 33.5±6.0 | -2.5±4.2 | 3.1±5.9 | -0.4±0.7 | -0.6±0.7 | 3.4±0.3 |
| AG | 3153 | 30.8±13.7 | -1.9±15.0 | 3.1±15.5 | 0.1±1.0 | -0.4±0.9 | 3.4±1.0 |
| CT | 17 | 29.7±5.9 | -0.4±4.0 | 5.7±4.9 | 0.4±0.4 | 0.3±0.8 | 3.4±0.3 |
| CT | 700 | 32.4±4.7 | -1.1±3.6 | 3.2±5.7 | 0.1±0.7 | -0.2±0.3 | 3.3±0.3 |
| CT | 1M | 33.5±6.0 | -2.5±4.2 | 3.1±5.9 | -0.4±0.7 | -0.6±0.7 | 3.4±0.3 |
| CT | 2632 | 32.3±9.7 | 1.3±12.2 | 2.3±12.9 | -0.0±0.9 | -0.2±0.9 | 3.4±0.7 |
| AT | 62 | 32.0±3.6 | 0.3±2.2 | -1.2±3.1 | 0.1±0.4 | -0.6±0.3 | 3.3±0.2 |
| AT | 890 | 30.0±4.7 | 0.2±2.9 | 1.6±5.3 | 0.0±0.5 | -0.6±0.4 | 3.2±0.3 |
| AT | 1M | 30.4±4.2 | 0.0±3.8 | -0.5±5.1 | 0.0±0.7 | -0.8±0.4 | 3.3±0.3 |
| AT | 3542 | 31.4±8.3 | -0.0±9.0 | -0.8±9.2 | -0.0±0.6 | -0.5±0.6 | 3.2±0.4 |
| CA | 30 | 41.9±8.7 | 0.7±2.7 | 0.7±7.0 | 0.1±0.2 | 1.6±1.0 | 3.3±0.2 |
| CA | 730 | 35.9±5.5 | 0.3±3.9 | 4.6±6.8 | -0.1±0.6 | 0.6±1.0 | 3.4±0.3 |
| CA | 1M | 29.6±8.3 | 0.2±4.8 | 10.3±6.3 | -0.2±0.7 | -0.2±0.6 | 3.1±0.4 |
| CA | 3013 | 34.5±13.1 | -0.4±10.6 | 4.6±12.3 | -0.0±1.0 | 0.2±1.3 | 3.4±0.9 |
| TG | 30 | 41.9±8.7 | 0.7±2.7 | 0.7±7.0 | 0.1±0.2 | 1.6±1.0 | 3.3±0.2 |
| TG | 730 | 35.9±5.5 | 0.3±3.9 | 4.6±6.8 | -0.1±0.6 | 0.6±1.0 | 3.4±0.3 |
| TG | 1M | 29.6±8.3 | 0.2±4.8 | 10.3±6.3 | -0.2±0.7 | -0.2±0.6 | 3.1±0.4 |
| TG | 2902 | 36.0±8.5 | -0.3±7.7 | 4.2±11.1 | -0.0±0.7 | 0.3±1.0 | 3.4±0.7 |
| CC | 36 | 33.8±5.6 | 1.9±3.4 | 4.3±4.6 | 0.0±0.5 | 0.7±1.0 | 3.4±0.3 |
| CC | 560 | 33.7±4.6 | 0.9±4.7 | 4.3±5.5 | 0.1±0.7 | -0.2±0.8 | 3.4±0.3 |
| CC | 1M | 32.7±6.6 | 0.0±4.4 | 4.9±5.7 | 0.2±0.7 | -0.7±0.9 | 3.5±0.3 |
| CC | 2718 | 32.2±12.5 | 1.4±9.8 | 3.0±10.8 | 0.1±0.8 | -0.2±0.9 | 3.3±0.7 |
| GG | 36 | 33.8±5.6 | 1.9±3.4 | 4.3±4.6 | 0.0±0.5 | 0.7±1.0 | 3.4±0.3 |
| GG | 560 | 33.7±4.6 | 0.9±4.7 | 4.3±5.5 | 0.1±0.7 | -0.2±0.8 | 3.4±0.3 |
| GG | 1M | 32.7±6.6 | 0.0±4.4 | 4.9±5.7 | 0.2±0.7 | -0.7±0.9 | 3.5±0.3 |
| GG | 2944 | 31.8±11.0 | -0.4±14.2 | 2.9±17.4 | -0.1±0.9 | -0.4±1.0 | 3.4±0.8 |
| CG | 205 | 33.0±5.1 | 0.3±4.5 | 3.4±7.7 | 0.1±0.5 | 0.5±0.6 | 3.3±0.3 |
| CG | 860 | 32.5±5.7 | -0.2±4.2 | 4.6±5.9 | 0.0±0.7 | 0.4±0.8 | 3.3±0.3 |
| CG | 1M | 29.8±8.8 | 0.0±5.2 | 9.3±6.0 | 0.0±0.7 | 0.0±0.5 | 3.1±0.4 |
| CG | 4283 | 30.7±15.4 | -0.9±14.2 | 7.4±17.6 | 0.0±0.8 | 0.4±1.6 | 3.4±1.2 |
| GA | 35 | 38.9±3.9 | -0.8±3.3 | 2.3±3.8 | 0.0±0.4 | 0.0±0.5 | 3.4±0.2 |
| GA | 805 | 35.4±4.7 | -1.4±3.6 | 2.3±5.5 | -0.2±0.6 | -0.1±0.6 | 3.3±0.2 |
| GA | 1M | 36.6±6.2 | -1.6±4.5 | 1.6±5.9 | -0.4±0.7 | -0.3±0.7 | 3.4±0.3 |
| GA | 3268 | 34.5±8.5 | -0.6±7.7 | 3.0±11.5 | -0.1±0.7 | -0.1±0.9 | 3.3±0.5 |
| TC | 35 | 38.9±3.9 | -0.8±3.3 | 2.3±3.8 | 0.0±0.4 | 0.0±0.5 | 3.4±0.2 |
| TC | 805 | 35.4±4.7 | -1.4±3.6 | 2.3±5.5 | -0.2±0.6 | -0.1±0.6 | 3.3±0.2 |
| TC | 1M | 36.6±6.2 | -1.6±4.5 | 1.6±5.9 | -0.4±0.7 | -0.3±0.7 | 3.4±0.3 |
| TC | 2847 | 34.7±9.6 | 1.1±7.4 | 3.3±9.4 | 0.2±0.7 | -0.0±0.8 | 3.3±0.5 |
| GC | 125 | 39.7±4.4 | -0.5±3.7 | -5.2±6.0 | -0.2±0.8 | 0.3±0.4 | 3.5±0.2 |
| GC | 940 | 35.0±5.2 | -0.2±3.7 | 0.6±5.7 | 0.0±0.7 | -0.1±0.6 | 3.3±0.3 |
| GC | 1M | 35.7±5.1 | 0.0±4.0 | -1.3±5.8 | 0.0±0.7 | -0.4±0.6 | 3.5±0.3 |
| GC | 4389 | 30.8±18.9 | 0.1±12.4 | 1.2±14.0 | -0.1±0.8 | -0.4±1.3 | 3.3±0.8 |
| TA | 19 | 41.7±6.3 | -0.1±3.5 | 1.4±5.0 | -0.2±0.4 | 0.7±0.8 | 3.4±0.2 |
| TA | 680 | 35.7±7.5 | 0.1±3.6 | 3.2±7.2 | 0.0±0.6 | 0.3±1.0 | 3.4±0.4 |
| TA | 25000 | 28.9±8.4 | 0.0±5.1 | 10.0±7.4 | 0.0±0.9 | -0.2±0.8 | 3.2±0.4 |
| TA | 3025 | 35.6±8.5 | 0.3±8.9 | 4.2±12.0 | 0.0±0.7 | 0.1±1.0 | 3.4±0.9 |
Units:
Translational Inter-BP Step Helical Parameters Shift/Slide/Rise in Å, Rotational Inter-BP Helical Parameters Tilt/Roll/Twist in degrees
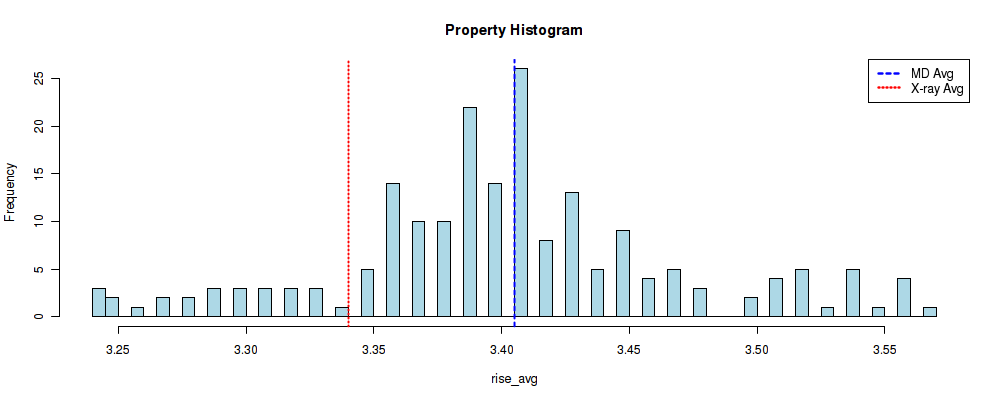
These computed experimental values are used as a comparison in global analyses plots (see analyses section). In the following example, vertical red line is showing the experimental average value found for the Rise helical parameter together with the histogram of Rise average values stored in the database for a particular base-pair step (in this case CG). Blue vertical line is showing the average value of the histogram data (MD simulations data).