Meta-trajectory Generation
BIGNASim allows the possibility to build meta-trajectories joining together nucleic acids fragments coming from different simulations. The generated trajectories can be downloaded or redirected directly to our NAFlex nucleic acid flexibility server to further analyse them.
The server will automatically show a button accompanied by an eye (Cassandra logo) when there is the possibility to generate a meta-trajectory, that is when working with a selected set of simulations containing a particular nucleic acid fragment.
Any particular fragment can be searched in the database using the search engine.
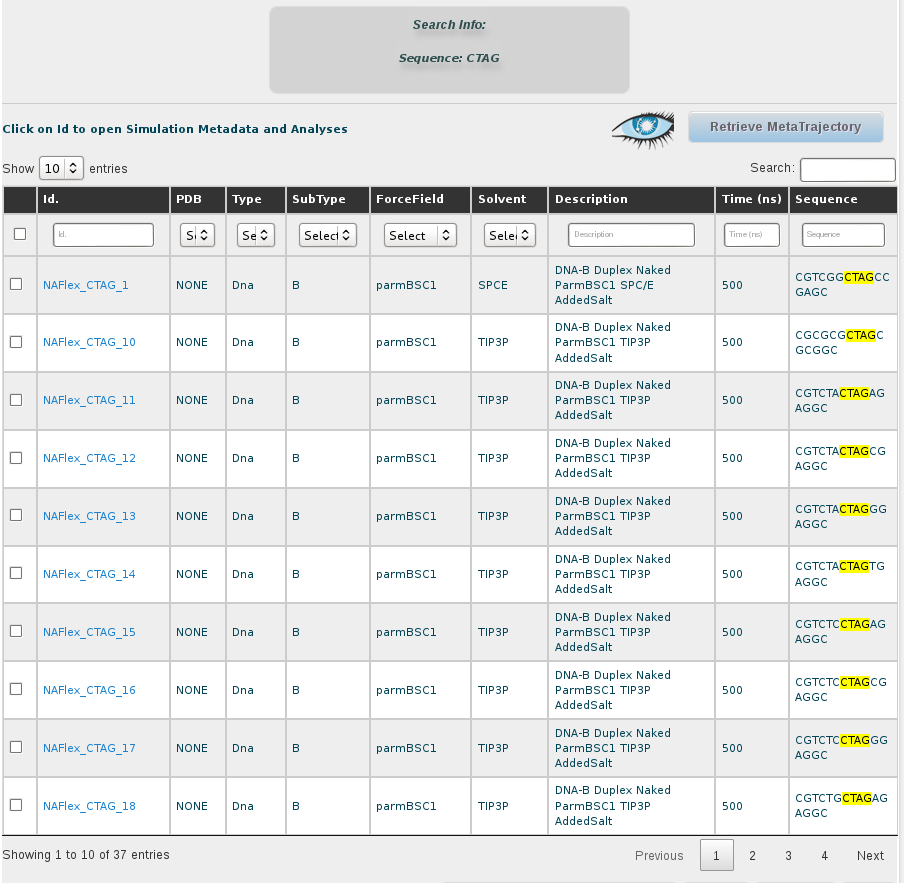
To retrieve a meta-trajectory, we must select a set of simulations from the ones shown in the browse section, just by clicking at the corresponding checkboxes placed in the left part of the screen.
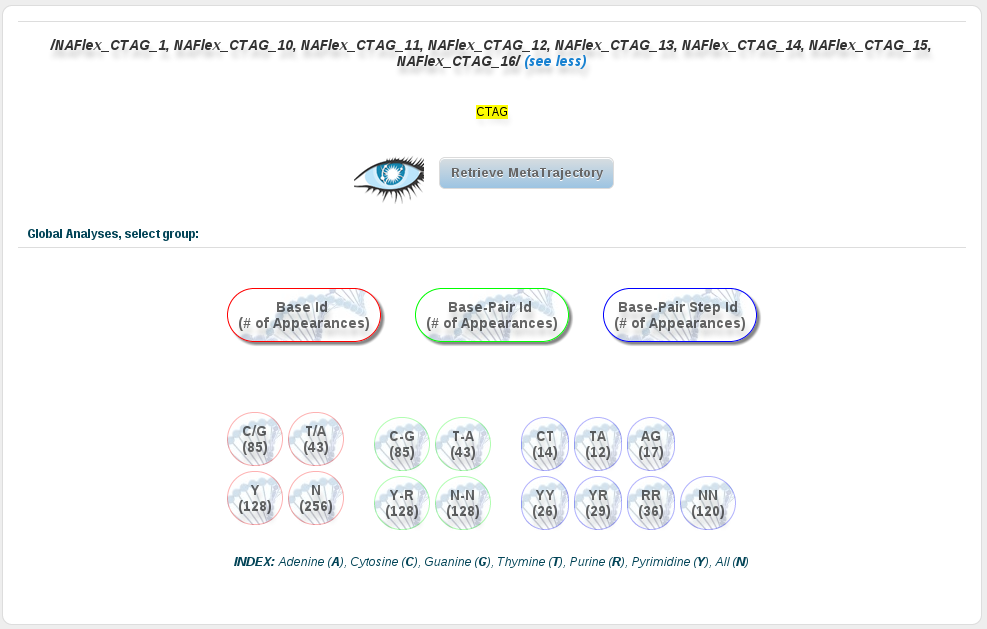
A fragment meta-trajectory can also be obtained from the global analyses section of the portal, as long as the global analyses we are looking at correspond to a particular nucleic acid fragment.
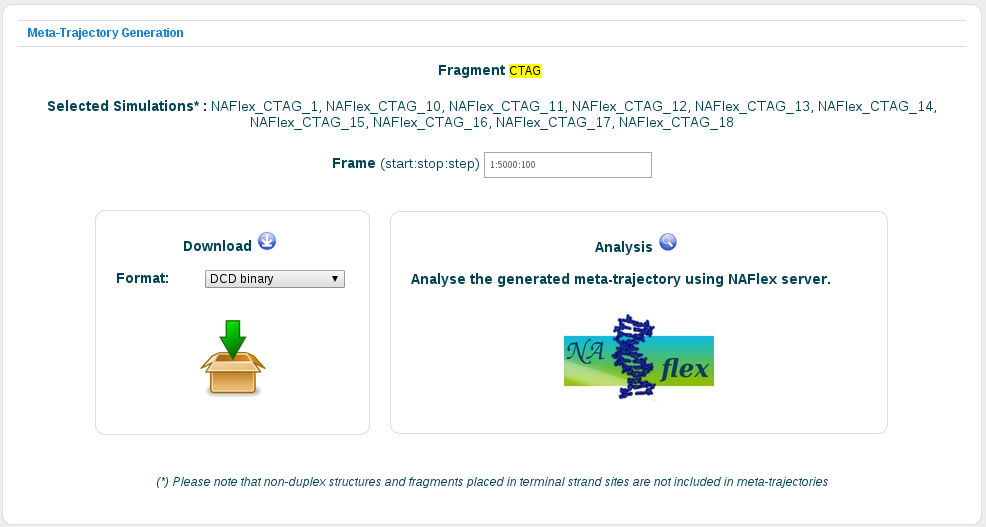
As a final stage before generating the meta-trajectory, BIGNASim offers a summary page, showing the desired nucleic acid fragment together with the complete list of trajectories selected.
In this same page, user could adjust the number of frames to be used from the original trajectories to generate the new meta-trajectory. Moreover, the newly generated trajectory can be downloaded or redirected to our NAFlex nucleic acid flexibility portal to further analyse it.