Bioactive Conformational Ensemble Help - REST API
REST API
Bioactive Conformational Ensemble DB provide programmatic access to the data through a REST API interface. The complete help section is accessible in this address. There, users have a list with all the available end-points distributed by different thematic blocks, all the models returned by these end-points and some practical examples of the REST API application.
End-points
- Generic Data: All the information available for each molecule
- Trajectory Analysis: Trajectory metadata for the requested molecule
- Trajectory Files: Structure and coordinate files generated for the requested molecule
- PCA Analysis: PCA data for the requested molecule
- PCA Files: PCA coordinates file for the requested molecule conformation
- MM Analysis: MM data for the requested molecule
- MM Files: MM coordinates and MIP files for the requested molecule
- QM Analysis: QM data for the requested molecule
- QM Files: QM coordinates for the requested molecule
- Libraries parameters: Parameters files for AMBER, CHARMM, CNS, GROMACS and OPLS
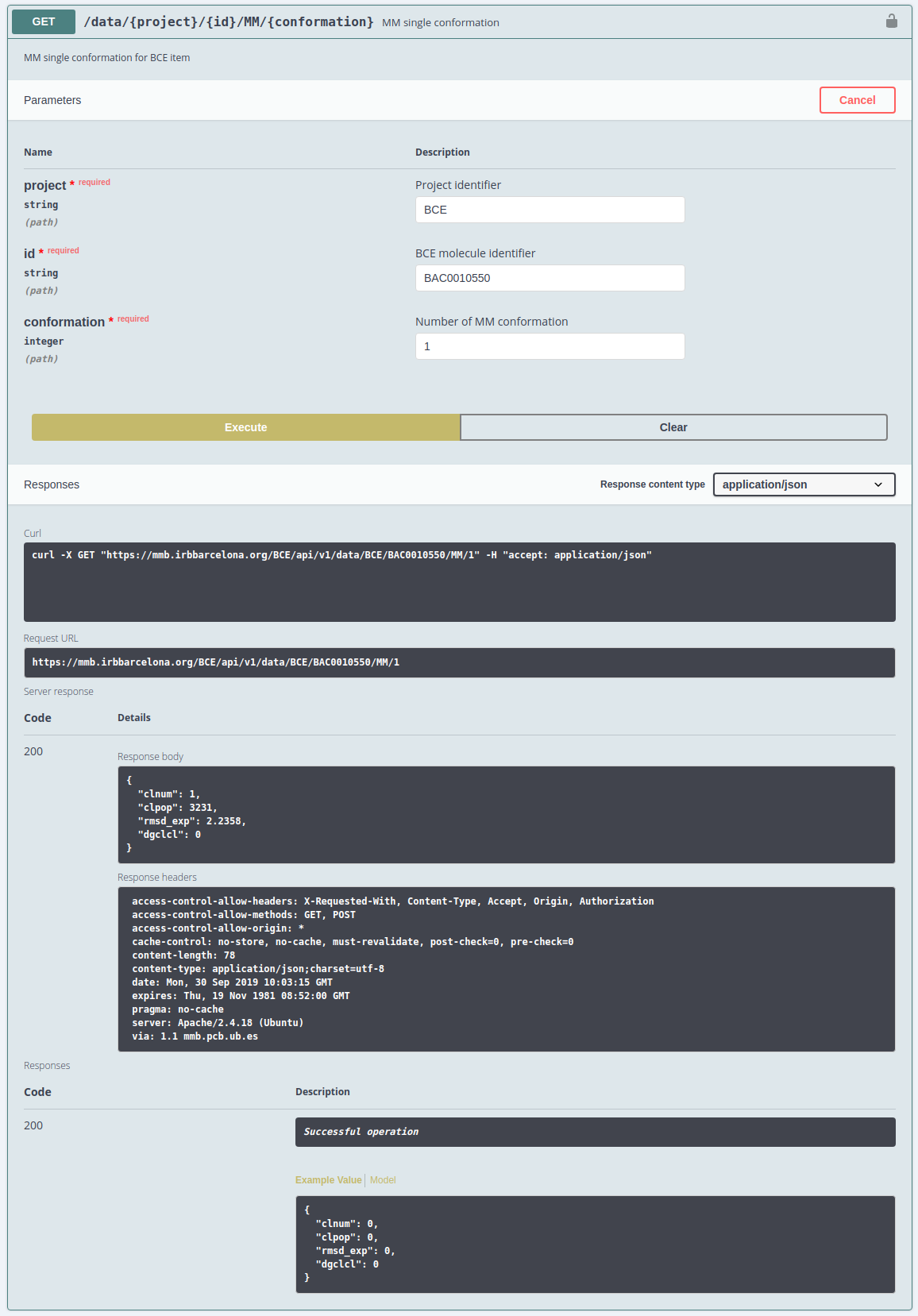
Clicking on each end-point, all the information needed for access it will be shown: parameters and possible responses. To get an example of the call, just click the top righ button Try it out and then the Execute big button and a cURL example call and its corresponding JSON response will be shown. Users can try to change the parameters and execute them.
Models
In this section all data models returned by the end-points are shown and described.
Practical examples
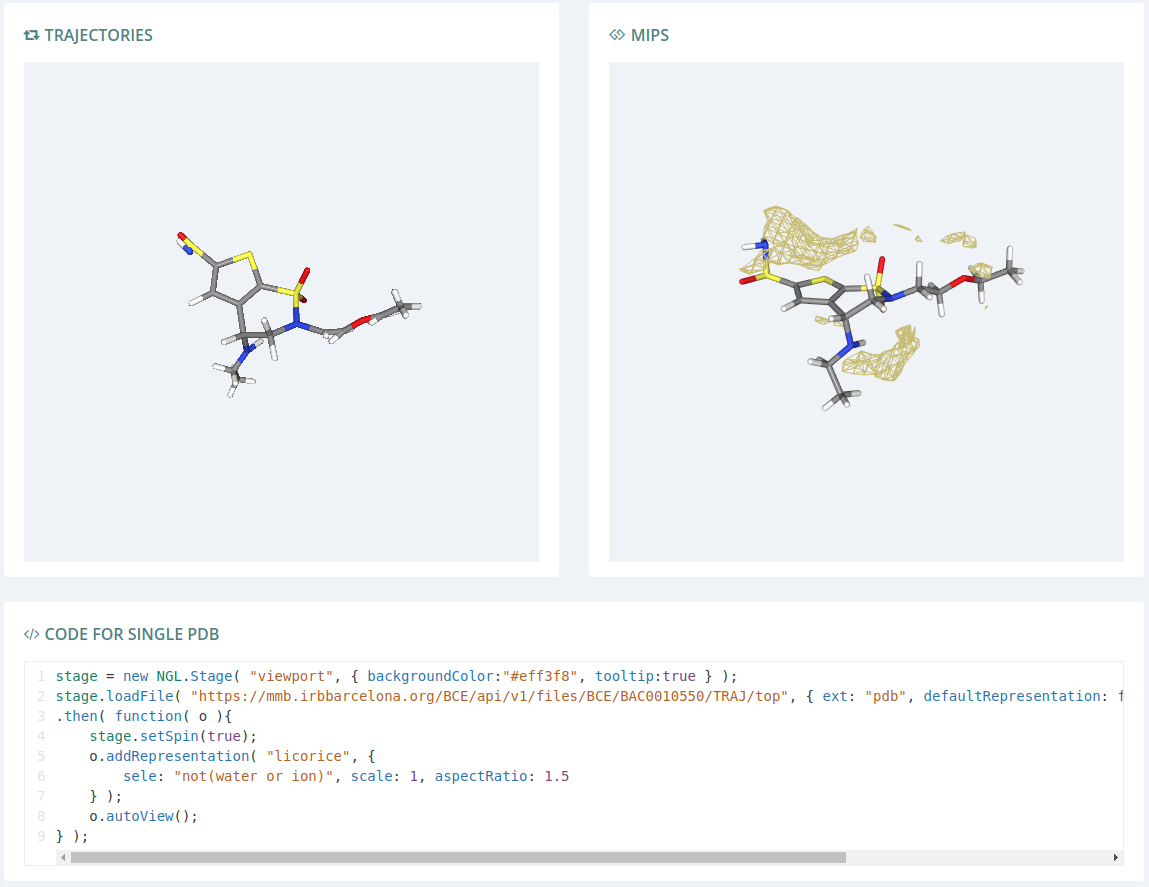
This section provides JS examples for calling the Bioactive Conformational Ensemble REST API and show the results via NGL Viewer. Three examples are provided:
- Single PDB: Load a structure
- Trajectories: Load a structure and its corresponding trajctory and animate them
- MIPs: Load a structure and its corresponding MIP file represented as a surface over the molecule