Bioactive Conformational Ensemble Help - Subsets Molecule
Molecule
The Molecule tab provides basic information about the small molecule, including general information, pharmacological properties and 2D/3D representations of the molecule, together with links to useful external websites. The module is divided in 4 components:
Please note that information related to the experimental structure will only be available if it has been experimentally solved previously (e.g. PDB code, protein-ligand complex visualization).
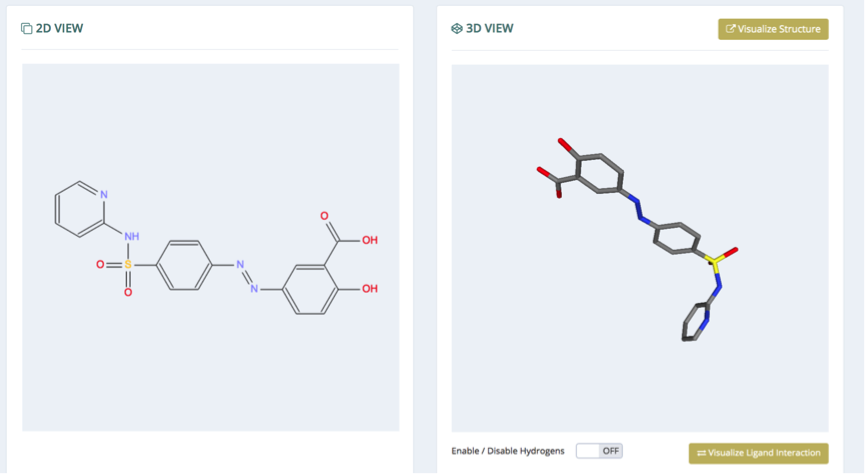
Molecule Information
General information about the molecule, in terms of chemical (formula, weight, chemical names & synonyms, InChI key) and structural (original PDB structure and ligand id, experiment type, resolution) evidences is shown in the first section. Links to the original websites from which data is retrieved (PDB, PubChem) are available in the bottom right part.
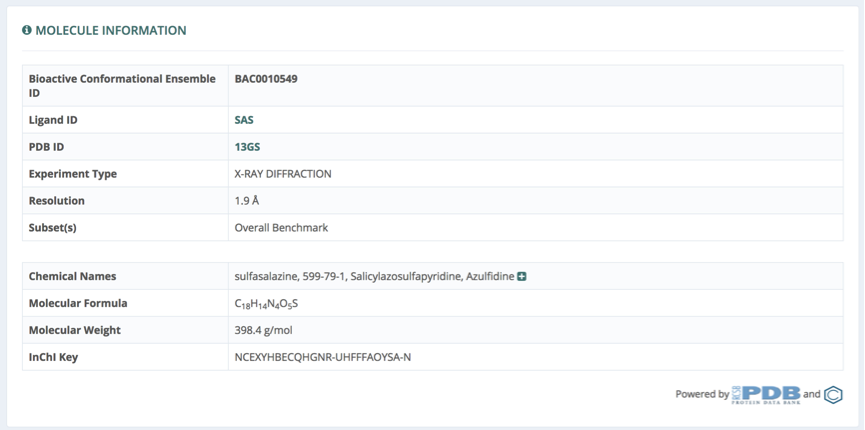
Molecule Properties
Pharmaceutical related properties of the small molecule are summarized in a table shown in the Molecule Properties section. Compliances of Lipinski and Veber’s rules evaluating molecule druglikeness are computed using the values from the table, and shown in two independent tables. Links to entries from a list of pharmacologically relevant libraries for the particular compound are available from a button in the top right part of the section. Databases linked will vary depending on the availability of the particular molecule as an entry in the database. Examples of such databases are: PubChem, DrugBank, SureChembl, Chembl, CheEBI, Therapeutic Targets Database (TTD), BindingDB and ZINC.
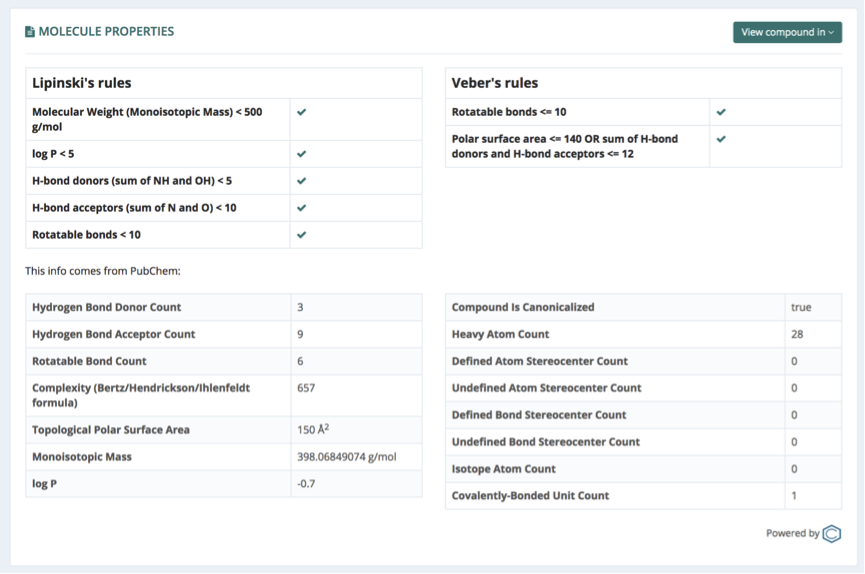
Smiles
Smiles section is showing the Simplified Molecular-Input Line-Entry System (SMILES) string code, generated through Open babel when the input ligand is in a different format (sdf, mol2 or pdb).
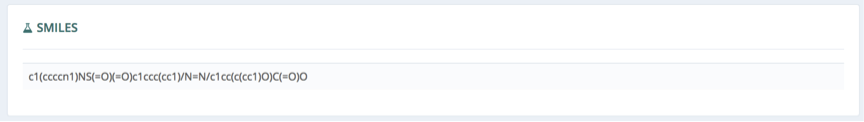
Visualization
Finally, in the visualization section, interactive 2D (left) and 3D (right) representations of the molecule are shown, with the possibility to show/hide hydrogen atoms, zoom in/out and rotate (3D). The Visualize Structure button opens a 3D representation of the protein-ligand complex system allowing a quick first look at the binding site position and shape. A second button (Visualize Ligand Interaction) opens a new browser tab with the RCSB PDB ligand interaction 3D view section, to further explore the experimental binding mode and the atoms associated in the protein-ligand interactions.