Bioactive Conformational Ensemble Help - Subsets Search
Browse / Search
Browse Database/Subset
Bioactive Conformational Ensemble DB allows you to browse the entire database or a particular database subset.
Menu: Bioactive subsets -> Overall Benchmark -> Browse Subset.

Information is presented in a graphical table. Each entry in the database is shown in a different table row, displaying the following information about the component (depending on availability):
- Entry ID, unique and persistent (FAIR) identifier for the database entry.
- 2D molecule structure.
- Drug name, including synonyms and alternative names.
- Molecular formula.
- Additional data, including PDB Code and Ligand ID, if an experimental complex is available, Lipinski’s and Veber’s rules fulfillment, and the particular database subset where the entry can be found.
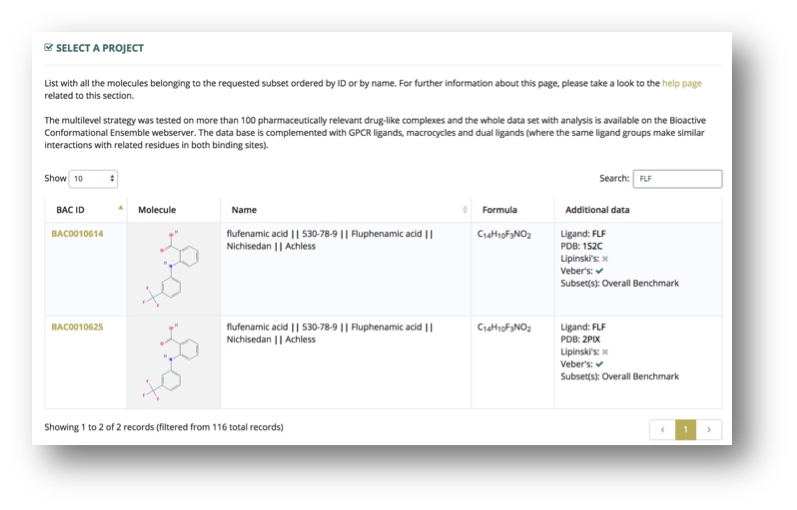
A search input box placed in the top-right part of the table allows quick searches by any type of textual information: name, alias, BAC id, ligand id, formula, PDB code, etc.
Clicking on one entry (table row) will redirect to the conformational ensemble information web section.
Search by Similarity
The web interface allows searching the database by molecular similarity.
Menu: Bioactive subsets -> Overall Benchmark -> Search by Similarity.

In the search by similarity section, a substructure can be introduced either from a SMILES code or from a drawn 2D representation using ChemAxon Marvin chemical editor. The engine is then searching the substructure within the database compounds using Tanimoto´s similarity search.

Search results are presented using the browse table interface (see Browse section), with an additional column showing the Tanimoto coefficient. The table allows the user to order the molecules by similarity according to this value, and also to search for important keywords such as BCE id, name, subset name or fulfillment of drug-like rules.
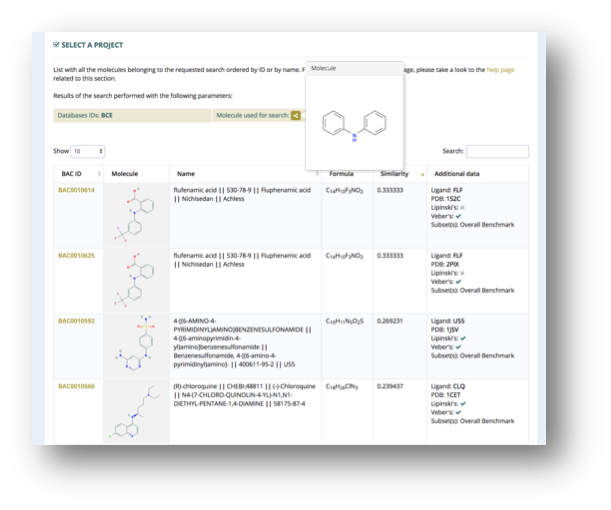
Clicking on one entry (table row) will redirect to the conformational ensemble information web section.
Search by Parameters
The web interface allows searching the database by different compound parameters: molecule description (e.g. name, id), molecule properties (e.g. number of heavy atoms, hydrogen bond donors/acceptors) and simulation results (e.g. ΔGdist , ΔGstrain).
Menu: Bioactive subsets -> Overall Benchmark -> Search by Parameters.

The possibility to filter those molecules fulfilling Lipinski’s or Veber’s rules is also integrated, which facilitates focusing the analysis in drug-like molecules.
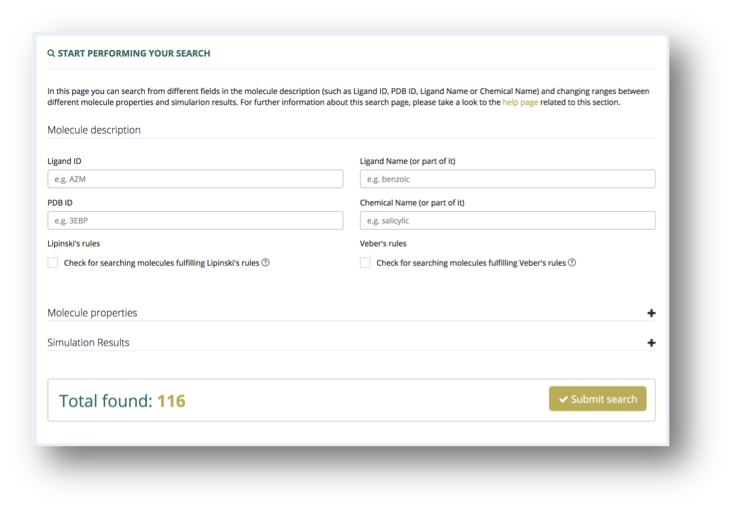
Search results are presented using the browse table interface (see Browse section), which also allows the user to search for important keywords such as BCE id, name, subset name or fulfillment of drug-like rules. The values ranges for the parameters used in the search are shown at the top of the page.

Clicking on one entry (table row) will redirect to the conformational ensemble information web section.
