Bioactive Conformational Ensemble Help - Create New Project - Browse / Search
Browse / Search
After some sections of the Step1, users will be redirected to the Browse Search page, where a list of matches (if there are matches) according to the parameters of search will be shown. In this list, users will be abe to select one molecule and, after visualize all its properties, start a project. This search of molecules can be performed through two databases: ChEMBL - EBI and Protein Data Bank.
ChEMBL - EBI
After selecting ChEMBL - EBI in the sections Chemical drawing, SMILES or Browse of the Step1: Upload data page, a search on the ChEMBL - EBI database will be performed (except in the cases where users have selected Create new project, in which case they will be redirected to the Step 2: Protonation state page).
Search Results
The search can be performed in two ways: by SMILES (the Chemical drawing section generates a SMILES code for the search) or by molecule name.
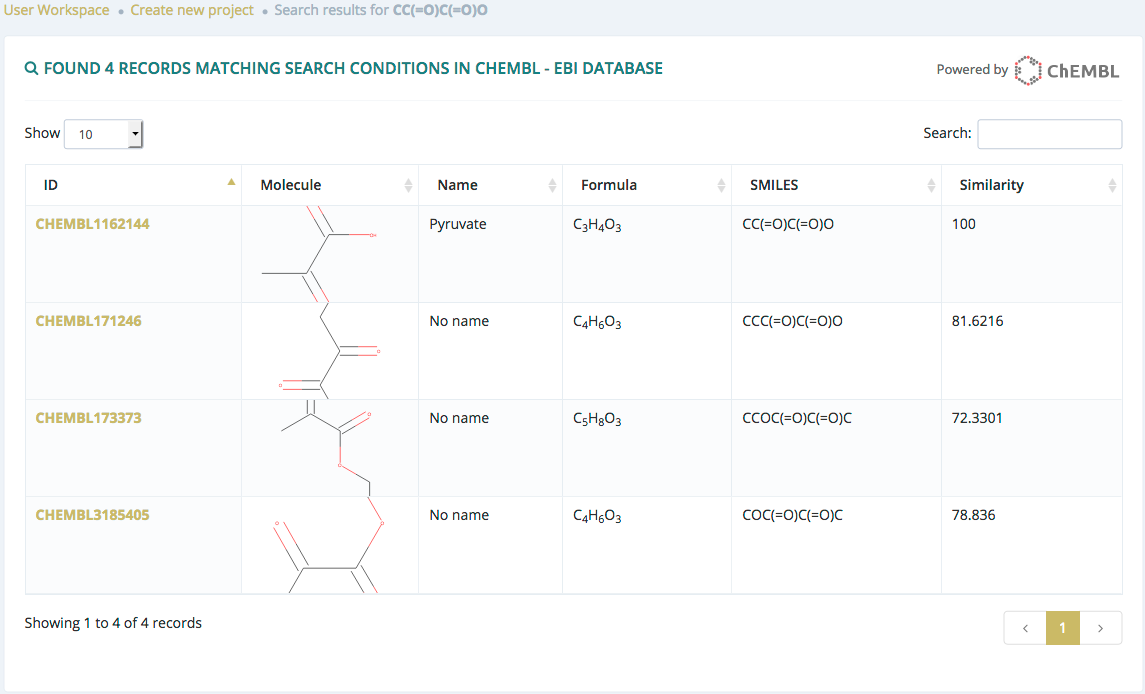
Search by SMILES
After submitting a search from the Chemical drawing or SMILES sections and if there are results, users will be redirected to a list of molecules.
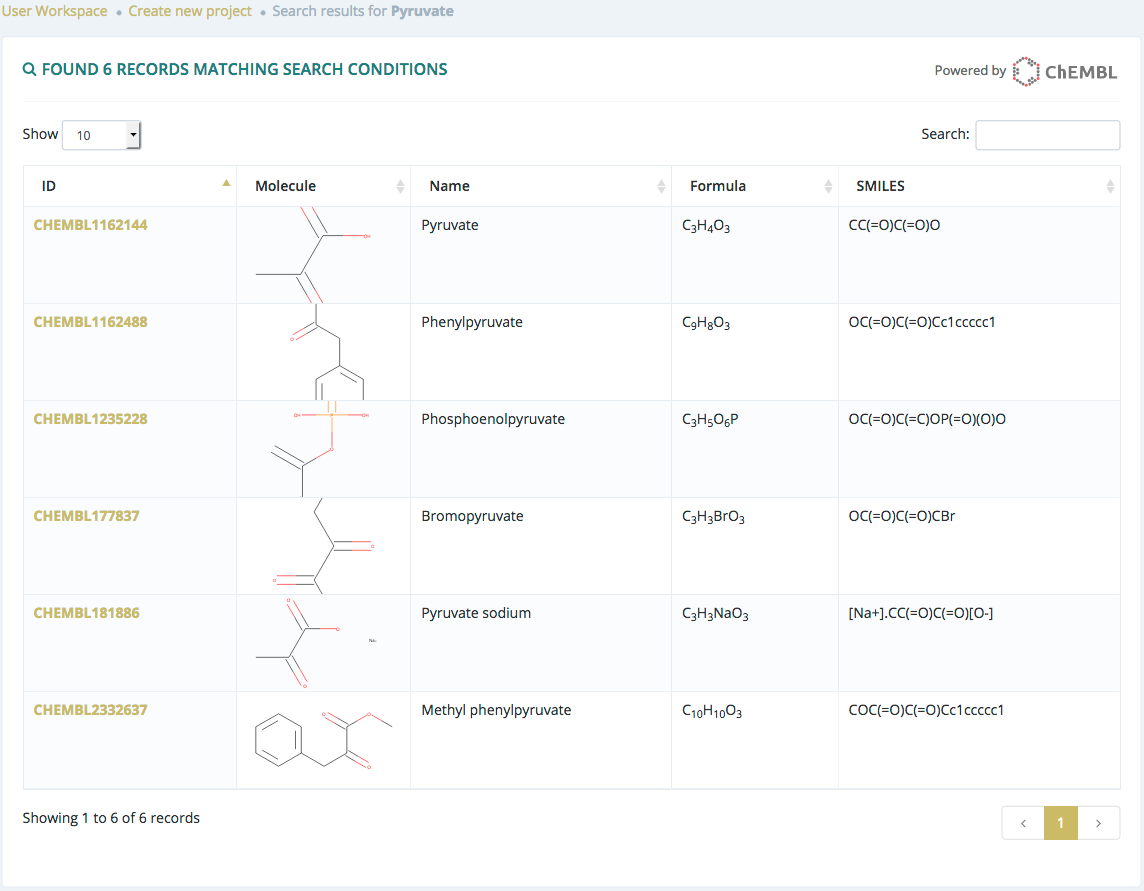
Search by molecule name
After submitting a search from the Browse section and if there are results, users will be redirected to a list of molecules.
Molecule metadata
After selecting a molecule from one of the lists above, users will be redirected to a page where is shown all the data and information associated to the molecule. For create a new project with this molecule, users just must click the Create new project with this molecule button. The molecule information is divided in four blocks:
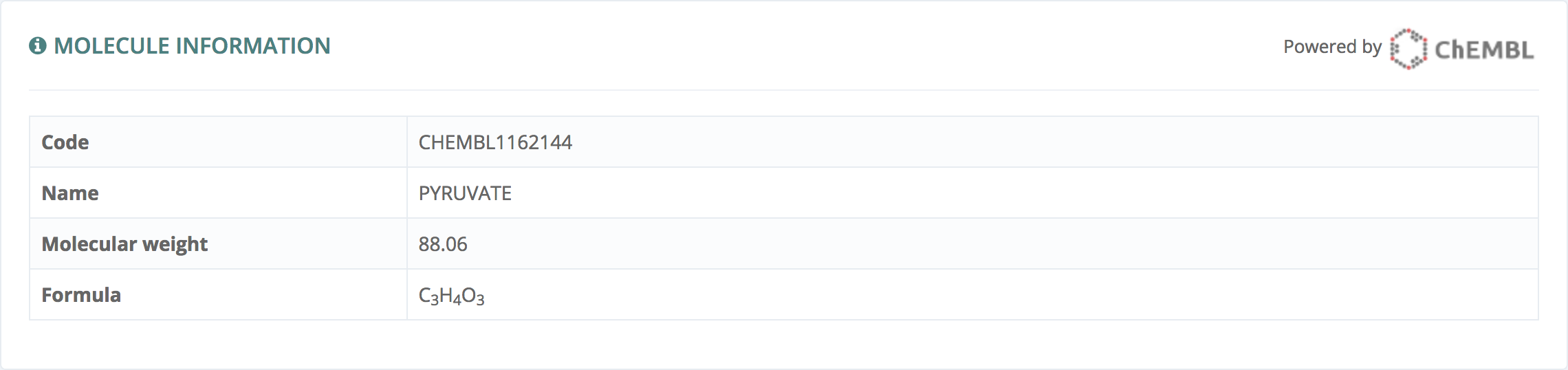
Molecule information
This information, powered by ChEMBL - EBI, gives ChEMBL code, Name, Molecular Weight and Formula.
Molecule properties
This information, powered by PubChem, tolds us if the molecule fulfills Lipinski's and Veber rules and gives several molecule properties.

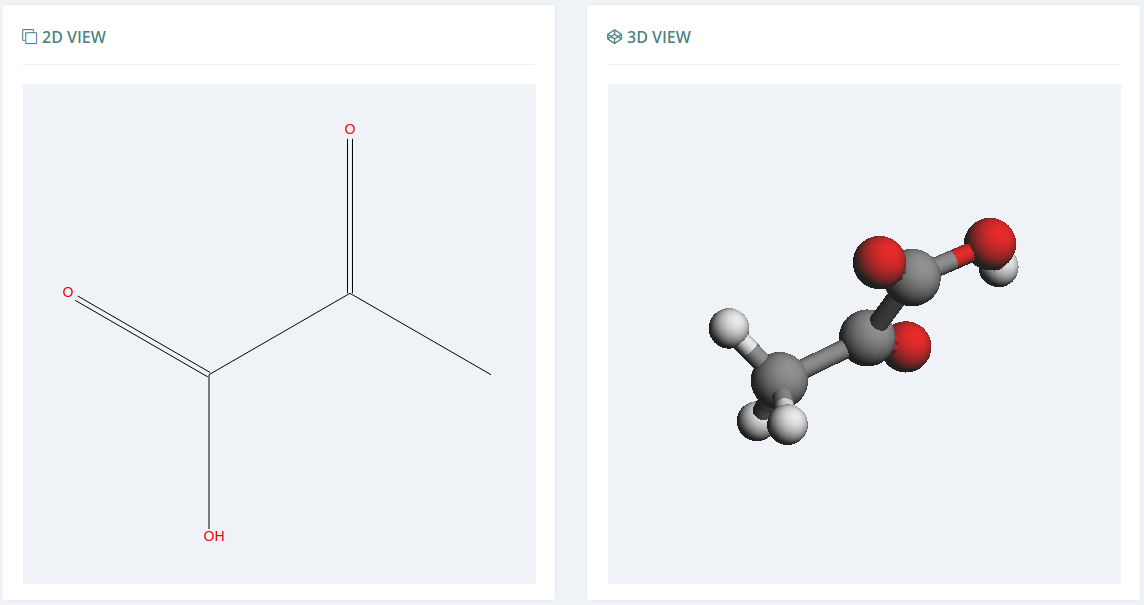
2D and 3D views
Through Open Babel and Kekule.js, the Bioactive Conformational Ensemble platform is able to build the 2D and the 3D views for the given molecule.
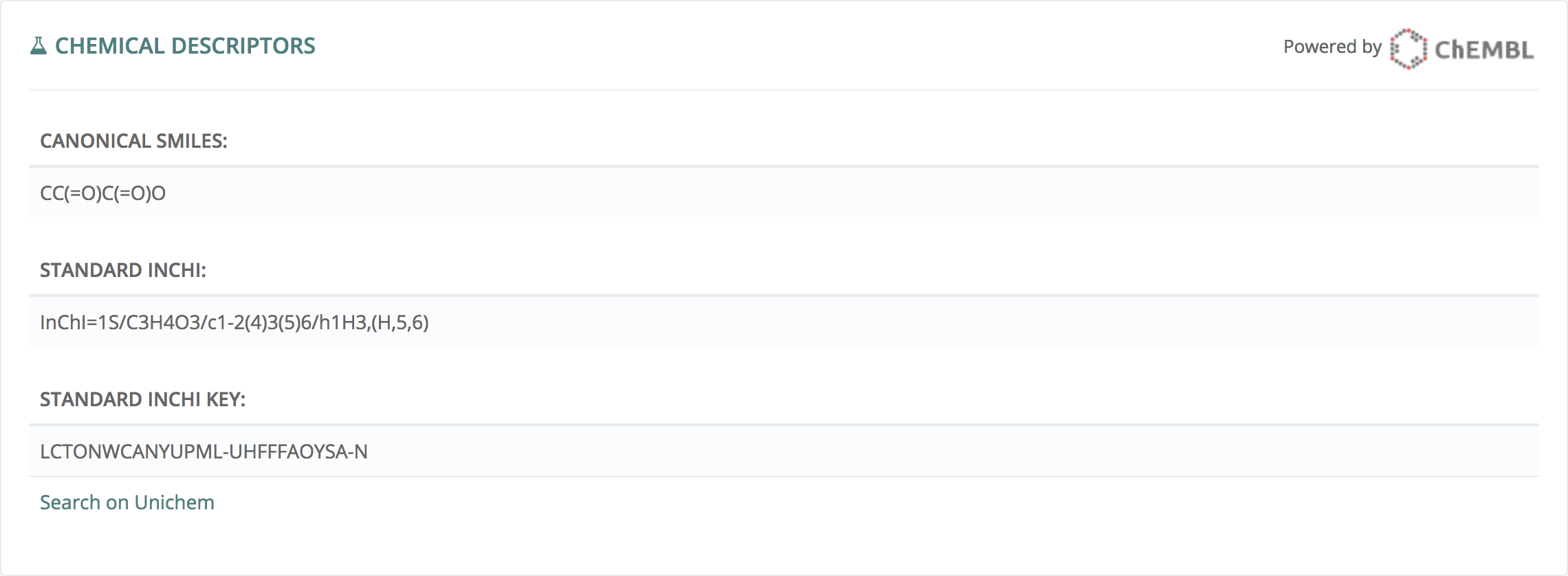
Chemical descriptors
This information, powered by ChEMBL - EBI, gives all the existing descriptors for this molecule.
Protein Data Bank
After selecting Protein Data Bank in the sections Chemical drawing, SMILES or Browse of the Step1: Upload data page, a search on the Protein Data Bank database will be performed (except in the cases where users have selected Create new project, in which case they will be redirected to the Step 2: Settings page).
Search Results
The search can be performed in two ways: by SMILES (the Chemical drawing section generates a SMILES code for the search) or by molecule name.
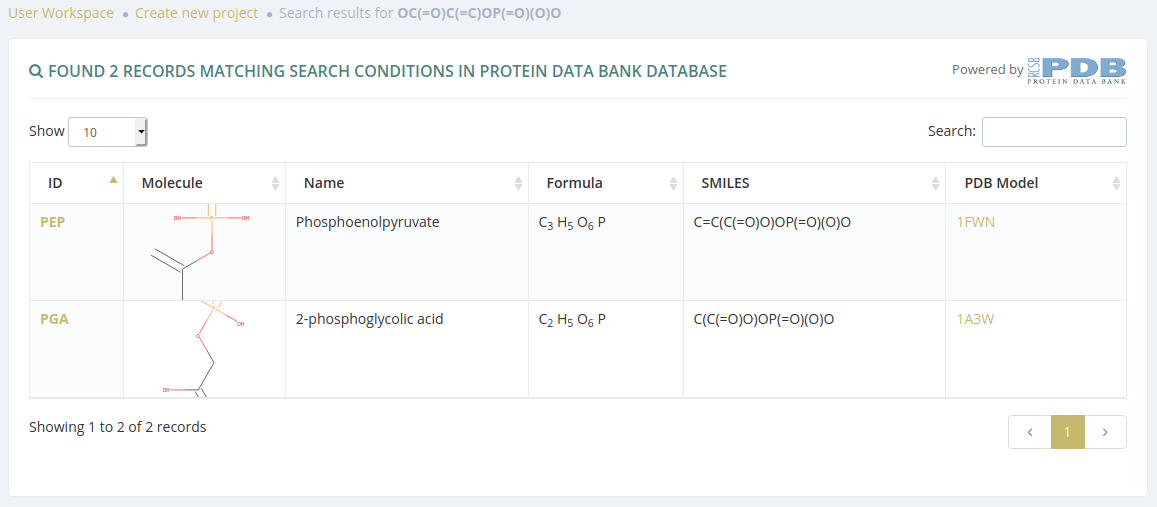
Search by SMILES
After submitting a search from the Chemical drawing or SMILES sections and if there are results, users will be redirected to a list of molecules.
Search by ligand name
After submitting a search from the Browse section selecting Ligand name and if there are results, users will be redirected to a list of ligands. Clicking the first column of the table, users will be redirected to Ligand metadata page, while clicking the last column, they will be redirected to Structure metadata page.

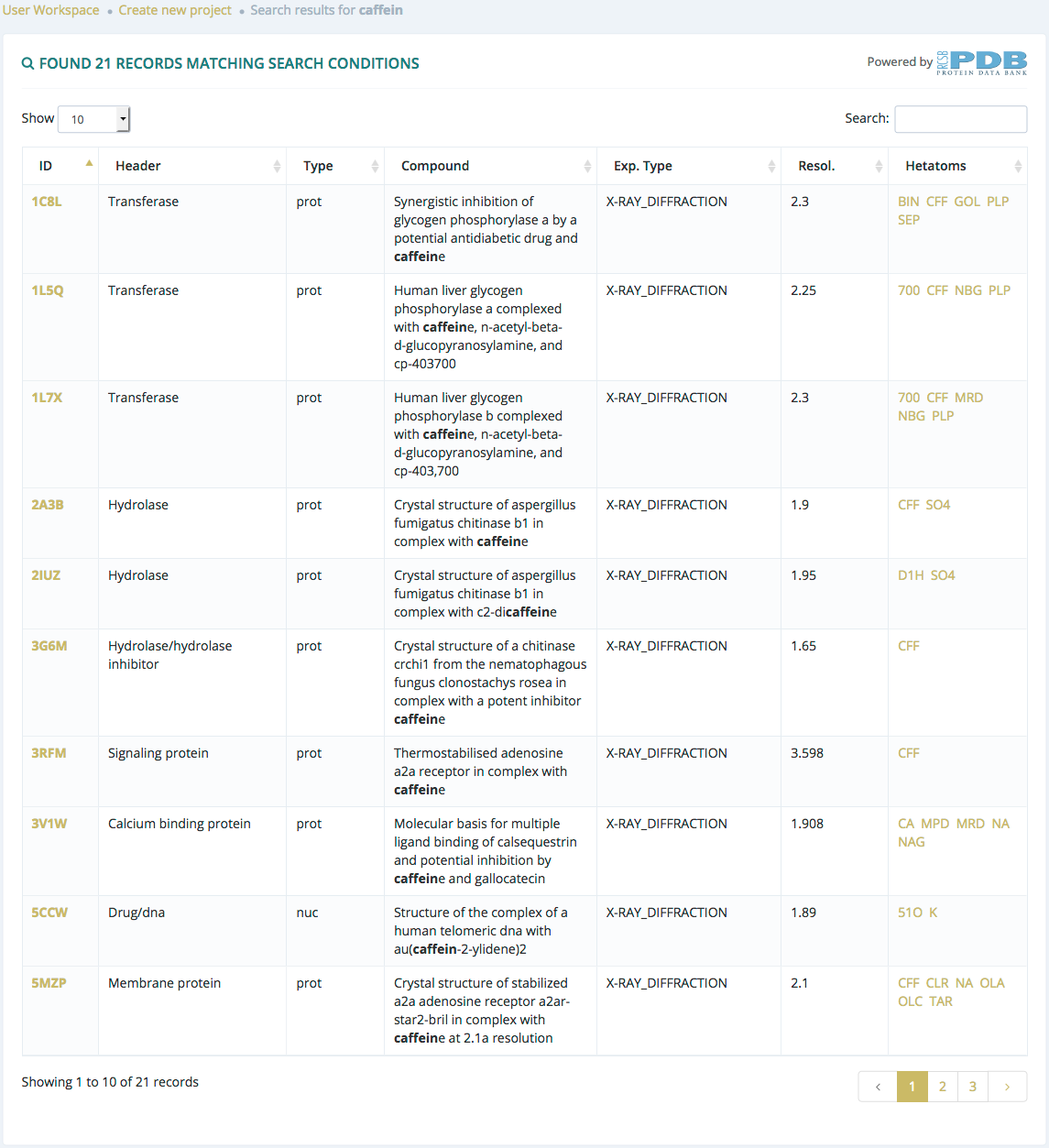
Search by structure header and compound
After submitting a search from the Browse section selecting Structure header and compound and if there are results, users will be redirected to a list of structures. Clicking the first column of the table, users will be redirected to Structure metadata page, while clicking the last column they will be redirected to Ligand metadata page.
Ligand metadata
After selecting a ligand from one of the lists above, users will be redirected to a page where is shown all the data and information associated to the ligand. For create a new project with this ligand, users just must click the Create new project with this ligand button. The ligand information is divided in six blocks:
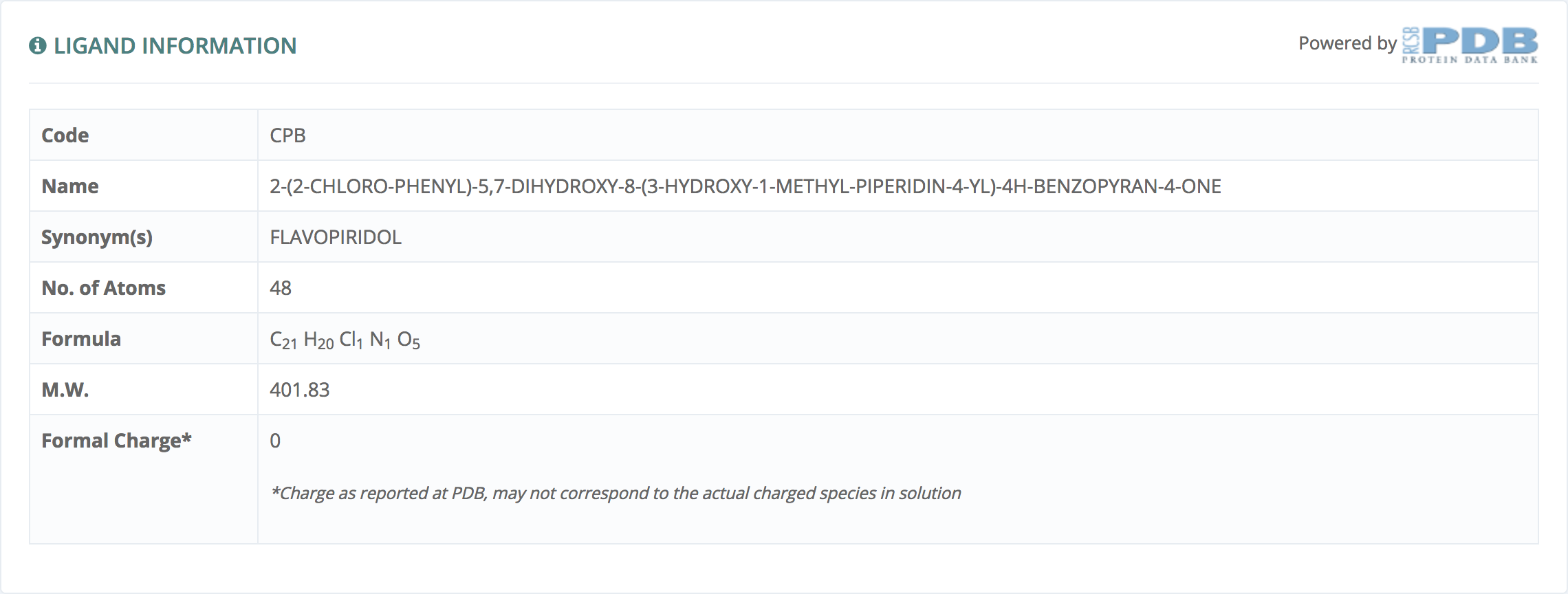
Ligand information
This information, powered by Protein Data Bank, gives PDB code, Name, Synonyms and all the relevant information provided by Protein Data Bank.
Molecule properties
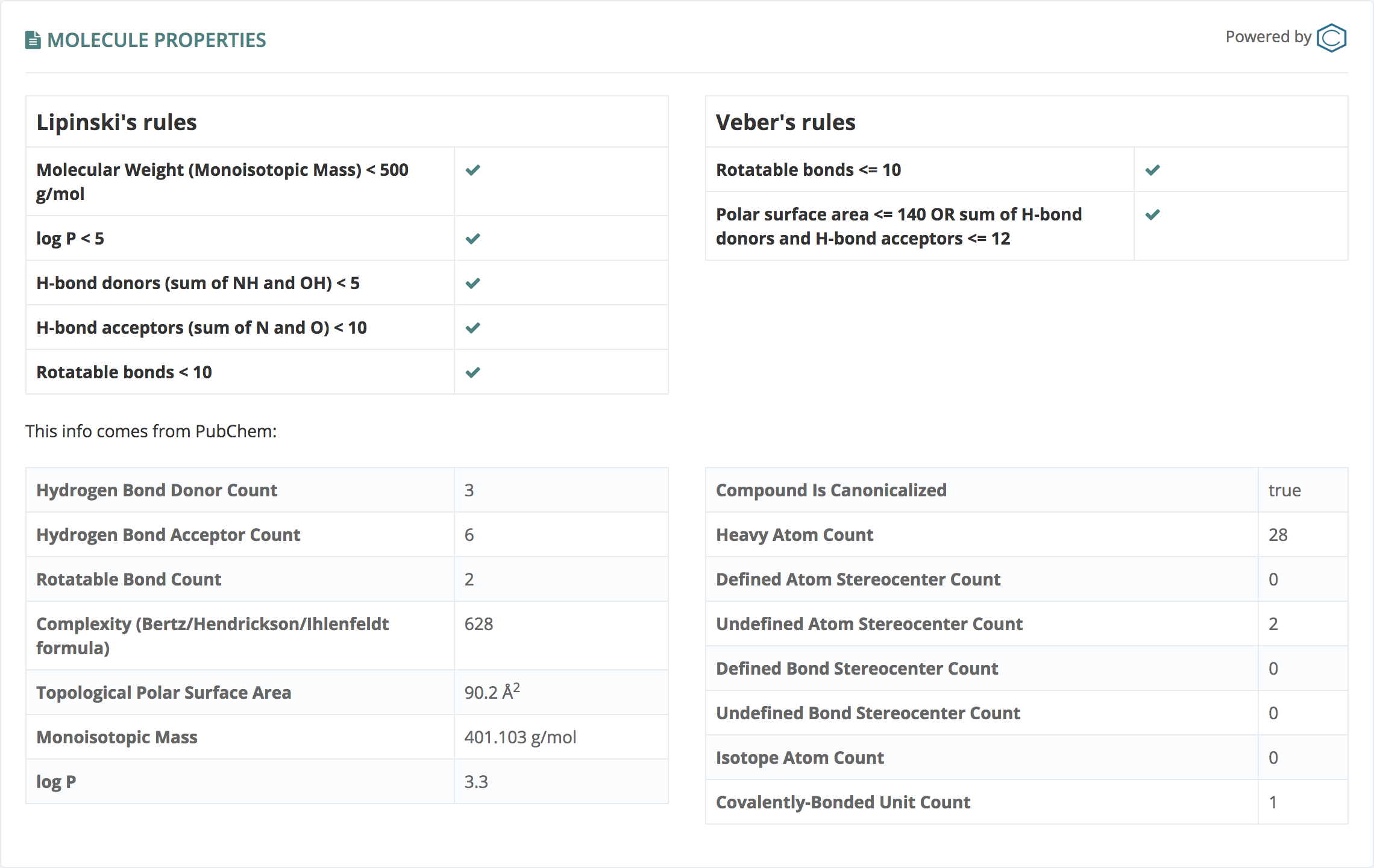
This information, powered by PubChem, tolds us if the molecule fulfills Lipinski's and Veber rules and gives several molecule properties.
2D and 3D views
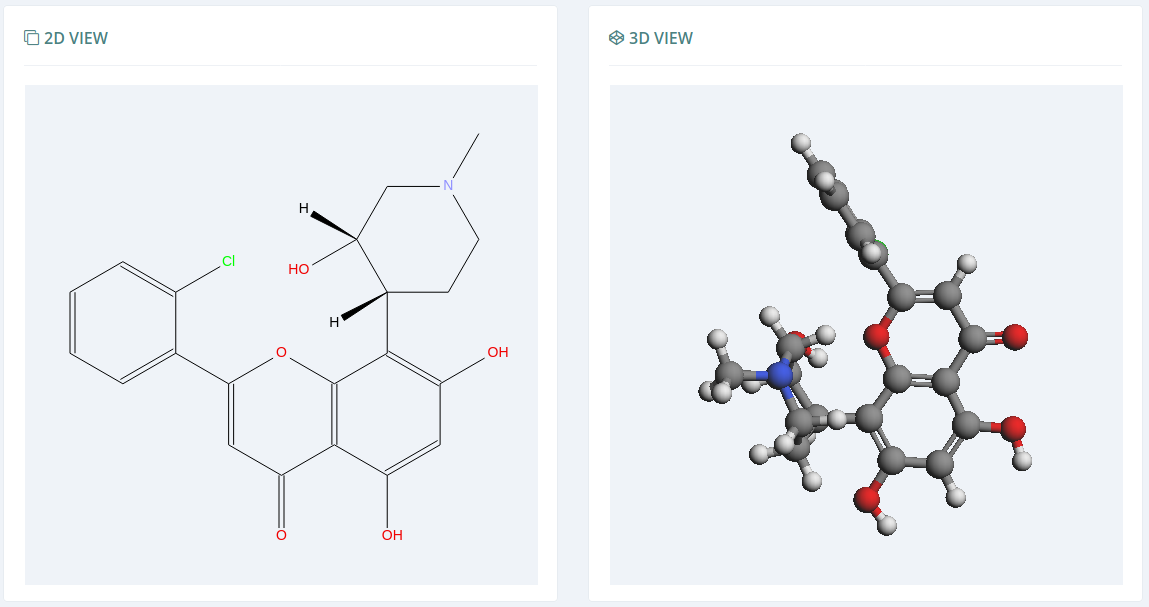
Through Open Babel and Kekule.js, the Bioactive Conformational Ensemble platform is able to build the 2D view for the given molecule. The 3D view is represented through NGL Viewer or Kekule.js (depending on the cases).
Systematic names
This information, powered by Protein Data Bank, gives the systematic names.

Chemical descriptors
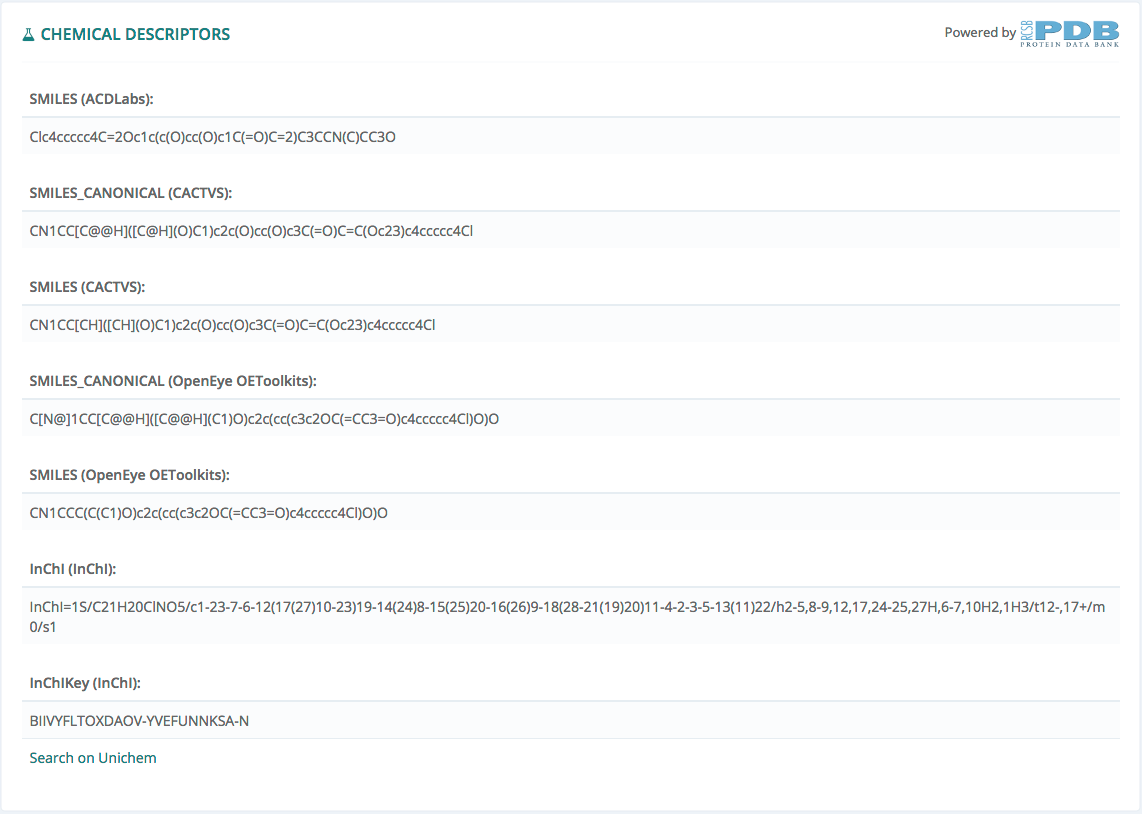
This information, powered by Protein Data Bank, gives all the existing descriptors for this ligand.
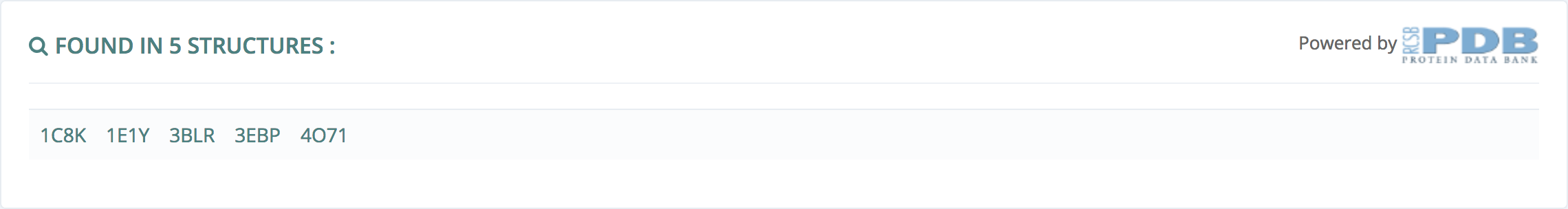
Found in N structures
This information, powered by Protein Data Bank, gives all the structures where this ligand can be found.
Structure metadata
After selecting a structure from one of the lists above, users will be redirected to a page where is shown all the data and information associated to the structure. For create a new project with a ligand contained inside this structure, users must go to the bottom of the page (LIGANDS section) and select one of them. The structure information is divided in four blocks:
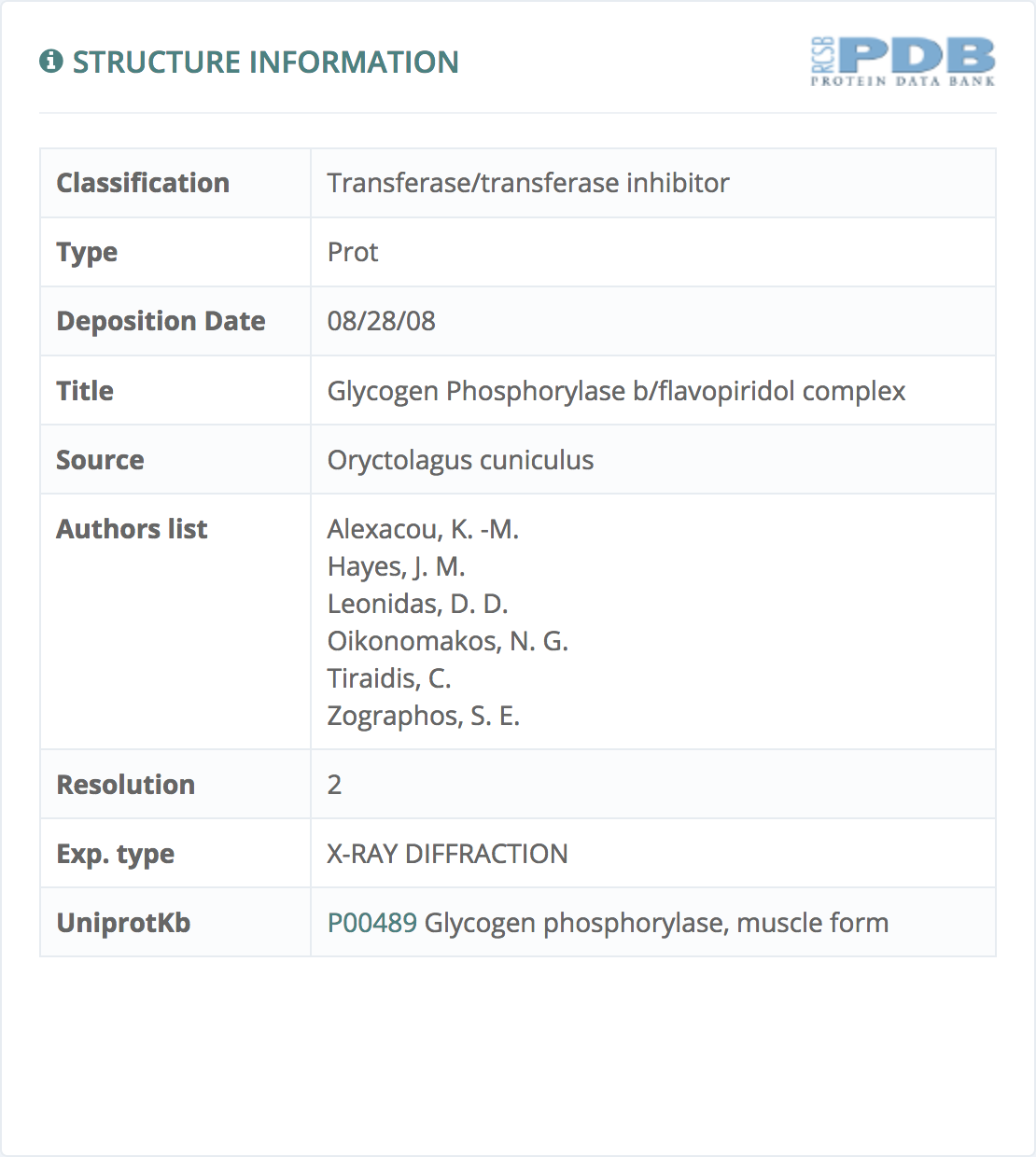
Structure information
This information, powered by Protein Data Bank, gives all the relevant information provided by Protein Data Bank.
3D view
The 3D view of the structure is represented through NGL Viewer .
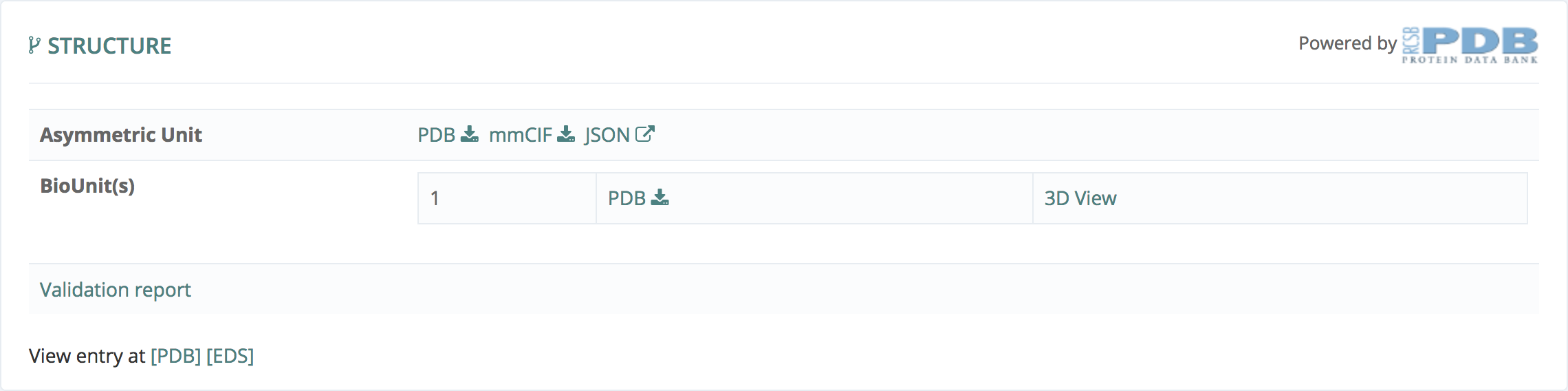
Structure
This information, powered by Protein Data Bank, gives all the information about Biounits and Asymmetric Unit.
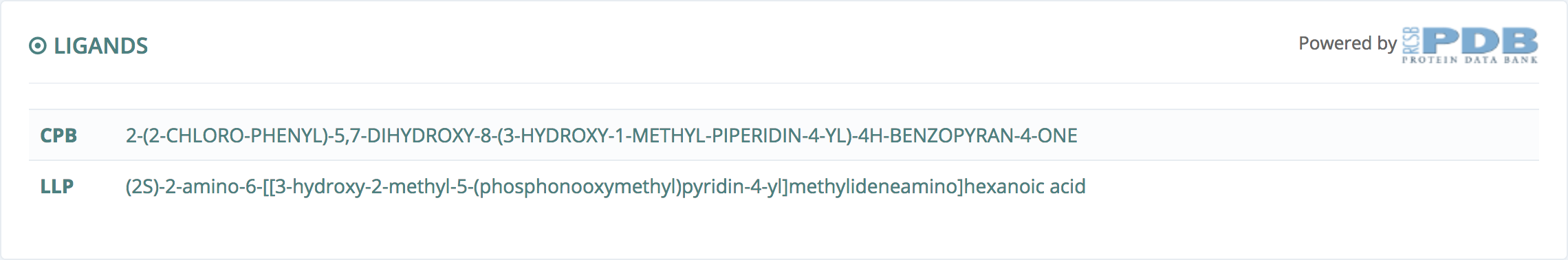
Ligands
This information, powered by Protein Data Bank, gives all the ligands found in this structure. For create a new project, users must select one of these ligands.