MC DNA Help - Inputs
Inputs Help
There are three groups of fields in the inputs page: generic fields, additional fields for Circular MC DNA and additional field for MC DNA + Proteins. All the fields have a icon. Rolling the mouse over this icon, a short help related to the field will be shown. Additionally, users can preload sample input data for launching jobs.
- Sample Input Data
- Generic fields
- Additional fields only Circular MC DNA
- Additional field only for MC DNA + Proteins
Sample Input Data
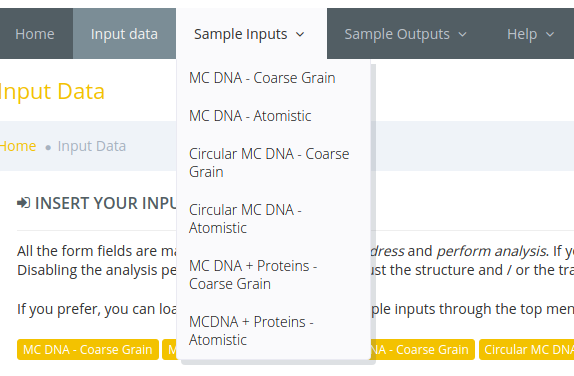
To load the sample input data, users should click one of the buttons of the Inputs page:

Also it's possible to access the sample input data via the main menu:
Generic fields
Write or paste DNA sequence
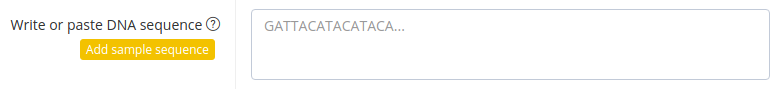
Put the full DNA sequence in this field. The sequence only may contain upper case letters A,C,G and T. The length of the sequence has to be between 15 and 500 nucleotides. No spaces or other signs are allowed in this field. Additionally, you can load a sample sequence clicking the left button "Add sample sequence".
Tool
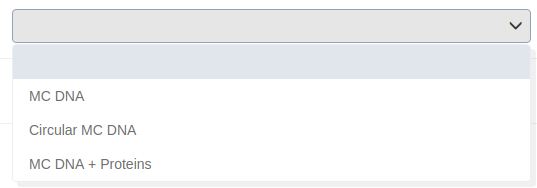
Select the tool you want to execute. You can choose between “MC DNA” which simulates naked B-DNA, “Circular MC-DNA” which simulates circular naked B-DNA and “MC DNA + proteins” which simulates the dynamics of B-DNA with proteins attached. See more details about the tools in “Methods”.
Resolution
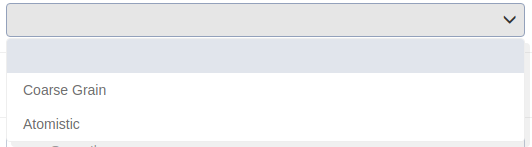
Choose the resolution of the selected tool. “Coarse Grain” resolution outputs a DNA structure with all atoms of the base pairs and the phosphate of the backbone while “Atomistic” has on top of the coarse grain structure all the backbone atoms included in the structure. All atoms of the DNA fiber are included. See more info in “Methods” under MC DNA.
Operations
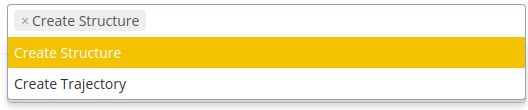
Two operations can be chosen. “Create Structure” creates a single structure with the ground state base pair step parameters of each base pair. “Create Trajectory” creates a certain number of DNA structures (specify the number of structures in the corresponding field “Number of structures”). Each of the structures is a final configuration after exhaustive Monte Carlo sampling on the bp-step parameters of the DNA. The option “Create Trajectory” is recommended if properties of the dynamics of the DNA want to be obtained.
Note: The range of analysis tools varies depending on the chosen type of operation (see “Analysis” for details)
E-mail address

Enter your E-mail address if you want to receive the link to the results once the simulation and analysis are done. This field is not mandatory.
Perform Analysis

Enable or disable flexibility analysis. Disabling this button the tool will only calculate the equilibrium structure and/or the trajectory and it will take less time.
Additional fields only Circular MC DNA
ΔLk

Choose a number of the change of the linking number ΔLk. Only integer numbers are valid. ΔLk = 0 means there will be no additional tension induced. Putting another number than 0 will induce over/undertwisting of the planar circular starting structure.
Iterations per structure

Choose the number of Monte Carlo iterations per structure. The higher the number the longer the simulation takes. In the following you can see a table formulas which should serve as a guideline for choosing the number of Monte Carlo iterations per structure for different ΔLk. The number of Monte Carlo iterations y depend on the size of the circle x (in bp). The formulas were fit to observations after how many Monte Carlo iterations the expected transition for a certain ΔLk is visible (for example the transition of a planar circle to a 8-shape for ΔLk=-1).
| ΔLk | Formula |
|---|---|
| 0 | No transition; y can be small (~ 1e6) |
| -1 | y = 67600 x + 2e07 |
| 2 | y = 303644 x – 3e-07 |
Circles of 94bp (Lankas et al., Structure 2006), 260bp (Noy et al., BJ 2017) and 336bp (Sutthibutpong et al, JCTC 2015) were used to fit these formulas. The value for required Monte Carlo iterations for ΔLk = -2 becomes negative for x ~ 100bp. This is because this formula was fitted to large minicircles with x > 260bp. Also take into account that the transitions may not occur because of the stiffness of the underlying sequence.
Additional field only for MC DNA + Proteins
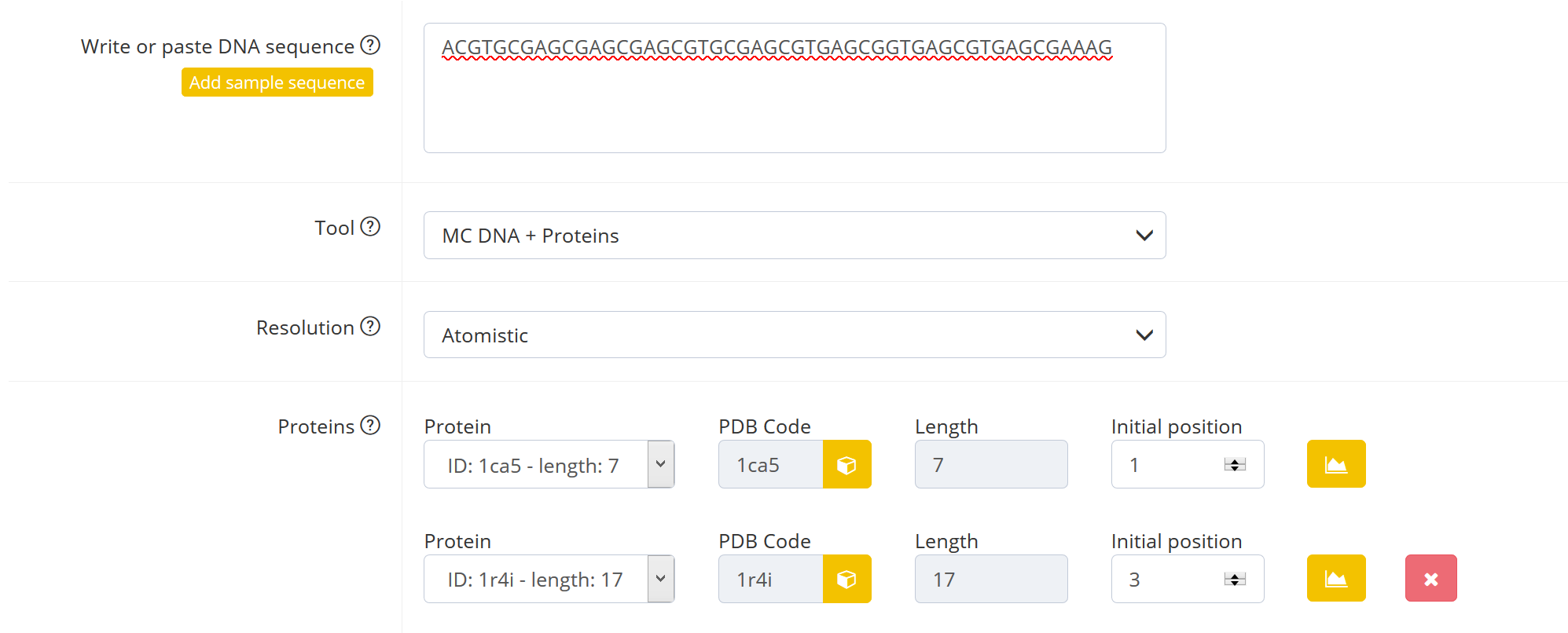
Proteins
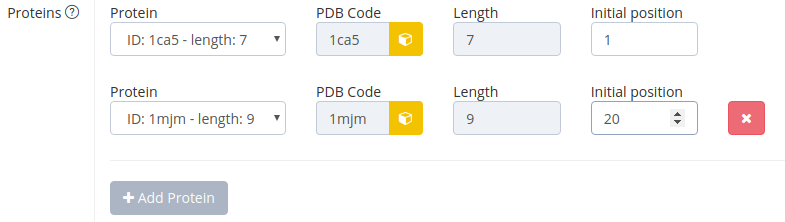
The user can select up to 10 proteins to bind to DNA. The ID is the PDB code and length is the number of base pairs bound to the protein. The user can define which base pair should be the first base pair bound to the protein in the field “Initial position”. Possible positions range from 1 to number of base pairs – number of base pairs bound to protein. For example the protein-DNA complex 1a0a is bound to 16 base pairs of DNA. If you choose initial position 2, the base pair steps 2 to 17 (which is base pair 3 to 18) are kept according to the base pair step parameters of the 1a0a complex. The sequence the user inputs for the bound DNA does not matter then.
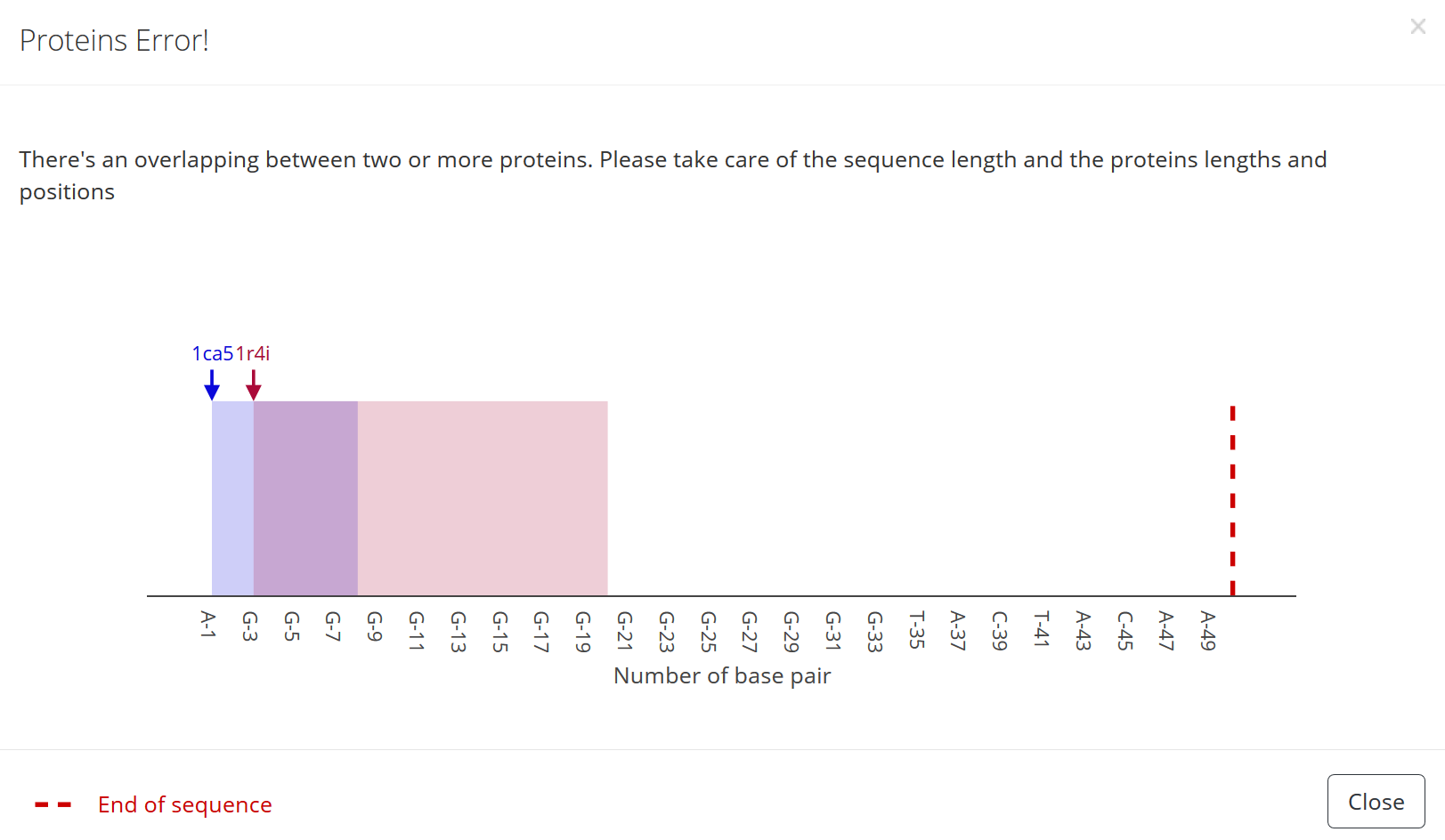
IMPORTANT: Make sure the bound DNA of one protein does not overlap with the bound DNA of another protein.
The user can also place the protein in an interactive way by clicking on the yellow graph button on the right.
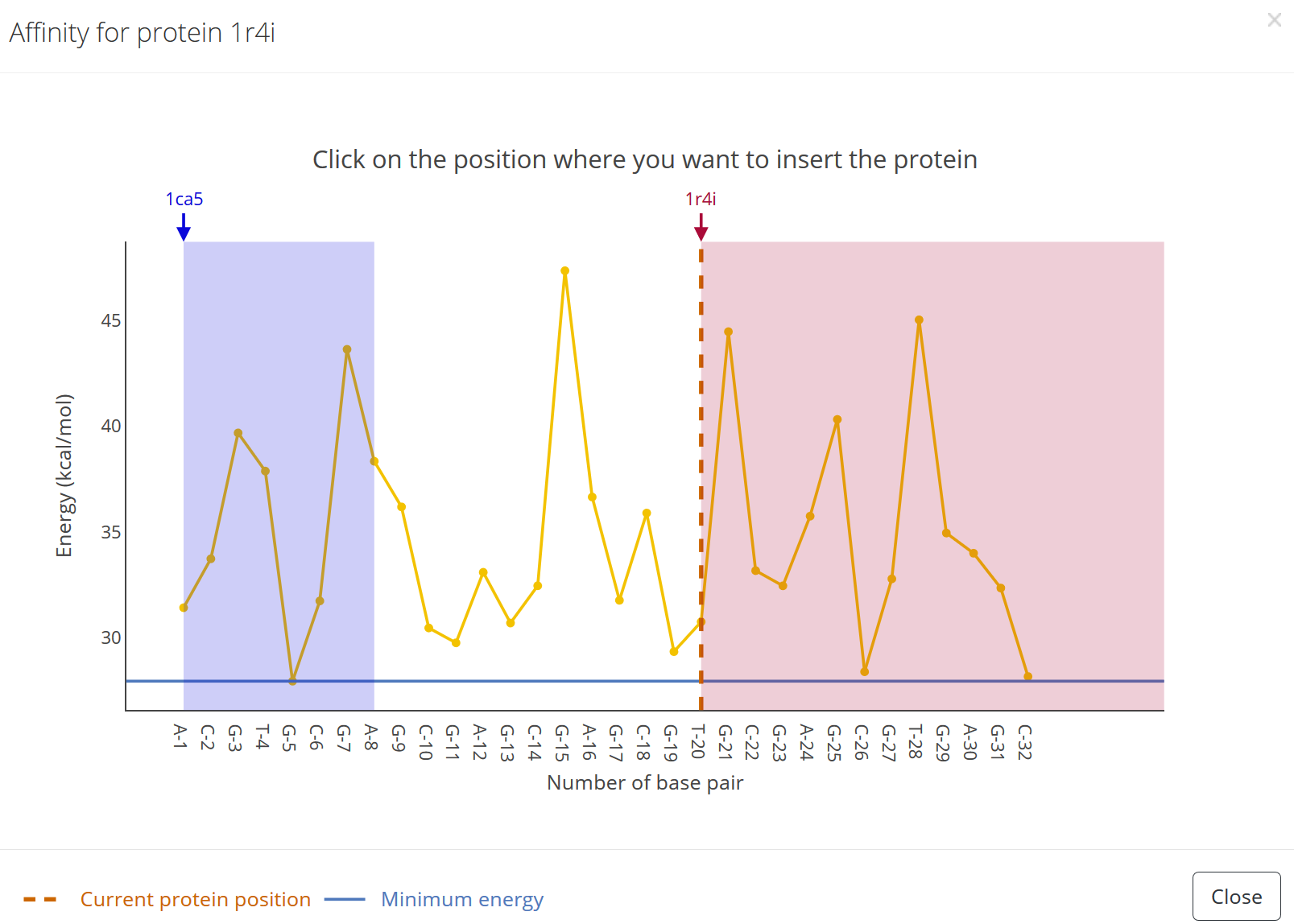
A graphical interface is shown where the DNA affinity for the proteins along the DNA sequence according to the propensity of the DNA to deform into the protein-bound DNA conformation derived from the pdb structure is plotted. The DNA occupied by proteins is colored. The dashed line shows the current position of the selected protein, the horizontal blue line shows the highest affinity/lowest elastic energy for the protein according to the deformability of the DNA. Within the affinity plot the user can select a new position for the protein by clicking on a data point in the graph. This position will be directly updated in the Input form.
Special cases:
1) In case the protein is positioned at the end of the sequence so that there are not enough nucleotides left to bind the protein an error message occurs once the “Submit” button is pressed:
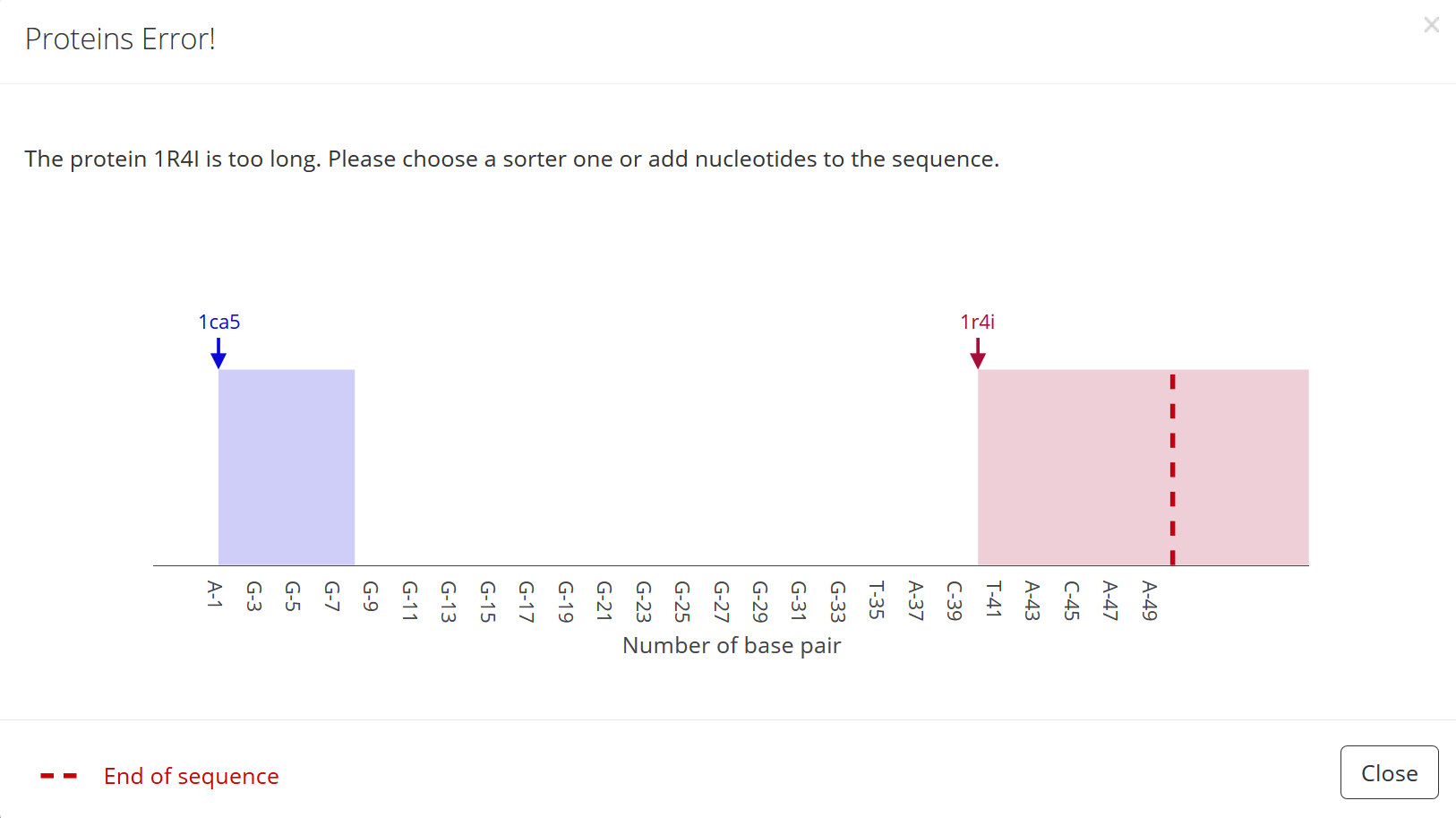
2) In a way that it overlaps with other DNA-bound proteins an error message occurs once the “Submit” button is pressed: