Help - Create project from Small Molecule
Create project from Small Molecule
In this section, users can launch a new project starting from a small molecule in order to parameterize it.
Provide Ligand
Upload PDB File
Upload a PDB file ligand from your computer with a maximum size of 10MB.
Once the file is selected, click the Submit button to proceed to the following step.
PDB ID + Ligand ID
For uploading the ligand from ID there are two choices: either selecting a ligand from a PDB ID the ligand by its ID.
When all the data is selected, click the Submit button step.
Browse
Perform a search on the Protein Data Bank. Write a text in the input box and the system will search this text in the header and the compound of each Protein Data Bank structure.
Once the results are shown, users can select by ligand ID.
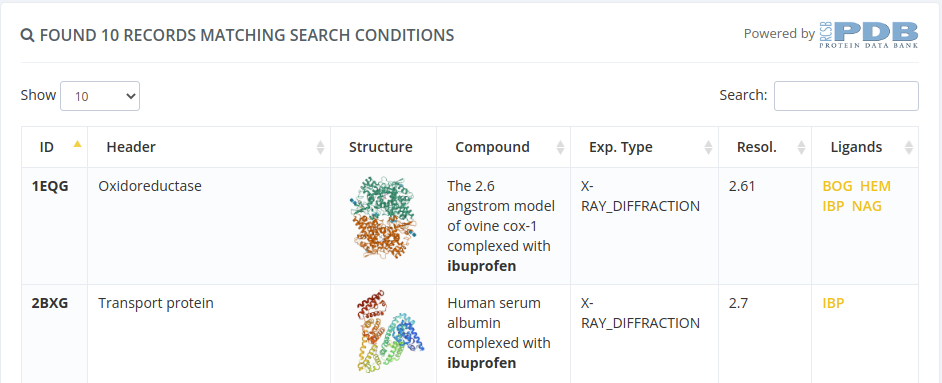
Selecting by ligand ID will open a page with all the properties of the parent structure of the selected ligand, but with a modal overlay with the ligand properties. Clicking in the Create new project button of this modal will proceed to the following step selecting this ligand.
Protonation state
If you want to preserve the original hydrogen atoms of the molecule and skip this step, just click the Preserve original hydrogen atoms of the molecule and skip this step button located at the top left corner of this page.
In order to compare the original molecule with the Major microspecies or the rest of the possible protonation states, you can click the Show original molecule button located at the top right corner of this page.
After clicking it, a new modal window with the original molecule will appear at the bottom of the page. This window can be dragged wherever you want and for closing it you just have to click the close button (located at the top right corner of the modal window).
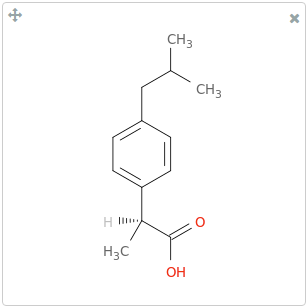
Major Microspecies
The Major Microspecies determines the major (de)protonated form of the molecule at a specified pH 7.4. In this page, the major microspecies is shown in the first place.
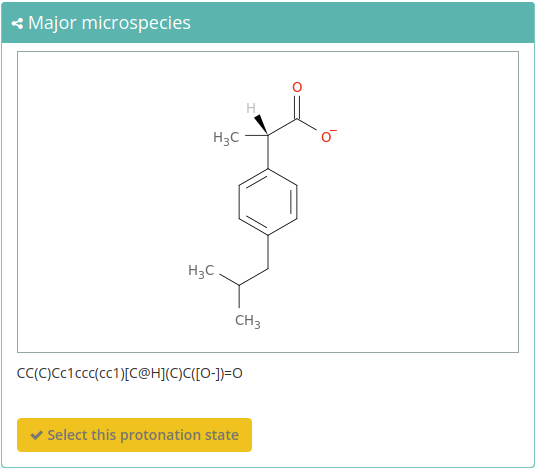
Possible Protonation states
Finally, clicking the Possible protonation states section at the bottom of the page, all the (de)protonated forms of the molecule at a specified pH in range from 0 to 14 will be shown:
For selecting the desired state you just need to click the Select this protonation state button in the bottom part of the cell.
Settings
In this section, users must provide a project name and, optionally, an email address for being notified once the workflow is finished. Take into account that some of the workflows can last several hours.
There are custom parameters for each workflow: when changing workflow, the custom workflow parameters change as well.
Workflows available if project has been created from Ligand:
GROMACS Ligand Parameterization
Click here for more information about this workflow.
GROMACS OPLS/AA Ligand Parameterization
Click here for more information about this workflow.
Antechamber Ligand Parameterization
Click here for more information about this workflow.
CNS/XPLOR Ligand Parameterization
Click here for more information about this workflow.
Run project
Before running the project, we can find a summary page with all the checking actions and workflow parameters, as well as a log of all the actions performed until this moment.
In this page, in the Download scripts section, users can download Python and YAML files for the sake of running the workflow at home. There are also available the CWL files for executing the workflow through Common Workflow Language. All the files have been customized with the parameters provided by users in previous steps.
Once all data is ok, we can click the Launch project button and the workflow starts showing its progress.
After finishing the running process, users will be automatically redirected to the output summary, broadly explained in the Outputs section.
